7X12
 
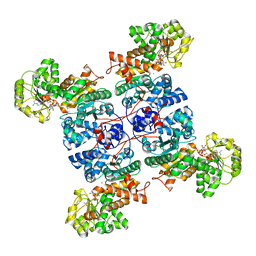 | | Crystal structure of ME1 in complex with NADPH | | Descriptor: | MANGANESE (II) ION, NADP-dependent malic enzyme, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and Characterization of a Novel Allosteric Small-Molecule Inhibitor of NADP + -Dependent Malic Enzyme 1.
Biochemistry, 61, 2022
|
|
7X11
 
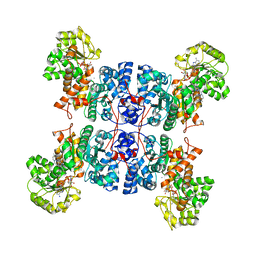 | | Crystal structure of ME1 in complex with NADPH | | Descriptor: | 6-[(7-methyl-2-propyl-imidazo[4,5-b]pyridin-4-yl)methyl]-2-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]-1,3-benzothiazole, MANGANESE (II) ION, NADP-dependent malic enzyme, ... | | Authors: | Amano, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery and Characterization of a Novel Allosteric Small-Molecule Inhibitor of NADP + -Dependent Malic Enzyme 1.
Biochemistry, 61, 2022
|
|
7FGL
 
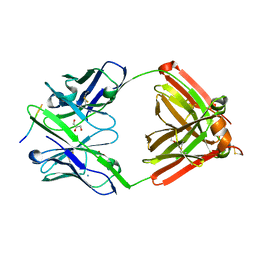 | | The complex structure of PHF core domain peptide of tau, VQIVYK, and antibody's Fab domain. | | Descriptor: | AMMONIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cross-reaction complex structure with VQIVYK of tau and the antibody's Fab domain.
To Be Published
|
|
2D3Q
 
 | |
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | Authors: | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
7YUZ
 
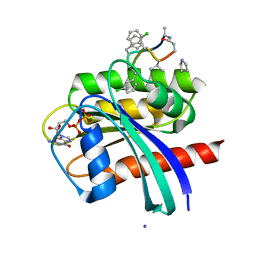 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP8784 | | Descriptor: | AP8784, GUANOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
6O3Z
 
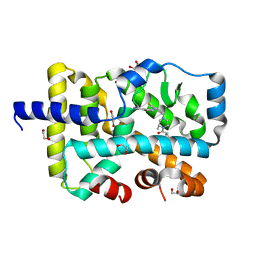 | | Crystal structure of RORgt with 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide (compound 1) | | Descriptor: | 1,2-ETHANEDIOL, 3-cyano-N-(3-{[(3S)-4-(cyclopentanecarbonyl)-3-methylpiperazin-1-yl]methyl}-5-fluoro-2-methylphenyl)benzamide, RAR-related orphan receptor C isoform a variant | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of [1,2,4]Triazolo[1,5-a]pyridine Derivatives as Potent and Orally Bioavailable ROR gamma t Inverse Agonists.
Acs Med.Chem.Lett., 11, 2020
|
|
1VOM
 
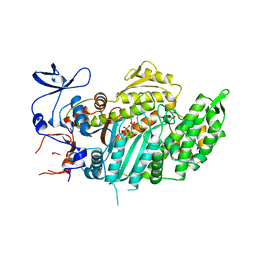 | |
1MND
 
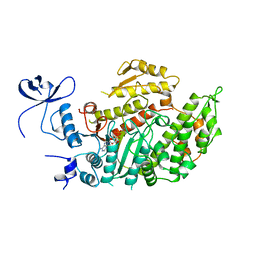 | |
1EQ3
 
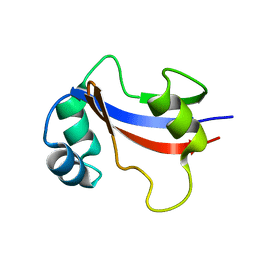 | | NMR STRUCTURE OF HUMAN PARVULIN HPAR14 | | Descriptor: | PEPTIDYL-PROLYL CIS/TRANS ISOMERASE (PPIASE) | | Authors: | Sekerina, E, Rahfeld, U.J, Muller, J, Fischer, G, Bayer, P. | | Deposit date: | 2000-04-02 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of hPar14 reveals similarity to the peptidyl prolyl cis/trans isomerase domain of the mitotic regulator hPin1 but indicates a different functionality of the protein.
J.Mol.Biol., 301, 2000
|
|
2GR2
 
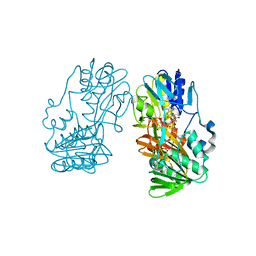 | |
6LE4
 
 | |
6LDO
 
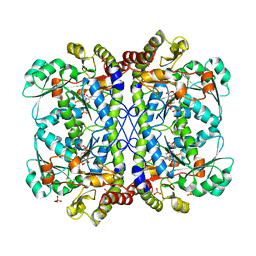 | |
8IGU
 
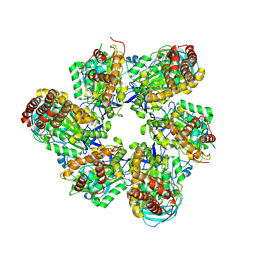 | |
8IGW
 
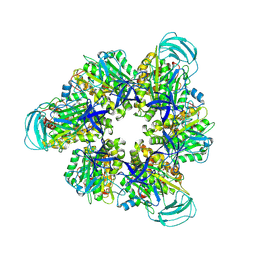 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
4X69
 
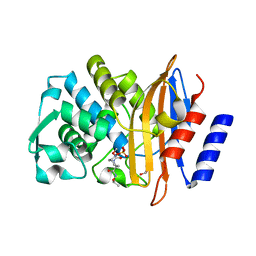 | | Crystal structure of OP0595 complexed with CTX-M-44 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase Toho-1 | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
4X68
 
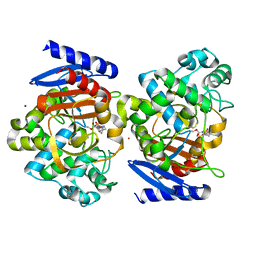 | | Crystal Structure of OP0595 complexed with AmpC | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, NICKEL (II) ION | | Authors: | Yamada, M, Watanabe, T. | | Deposit date: | 2014-12-07 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | OP0595, a new diazabicyclooctane: mode of action as a serine beta-lactamase inhibitor, antibiotic and beta-lactam 'enhancer'
J.Antimicrob.Chemother., 70, 2015
|
|
3VHW
 
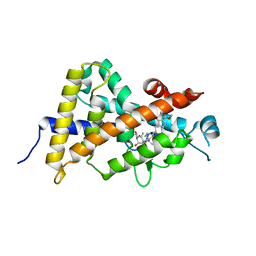 | | Crystal structure of the human vitamin D receptor ligand binding domain complexed with 4-MP | | Descriptor: | Vitamin D3 receptor, methyl (1S,3E)-3-{(2R)-2-[(1R,3aS,4E,7aR)-4-{(2Z)-2-[(3R,4S,5R)-3,5-dihydroxy-4-(3-hydroxypropoxy)-2-methylidenecyclohexylidene]ethylidene}-7a-methyloctahydro-1H-inden-1-yl]propylidene}-1-ethyl-2-oxocyclopentanecarboxylate (non-preferred name) | | Authors: | Kakuda, S, Takimoto-Kamimura, M. | | Deposit date: | 2011-09-08 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Synthesis of novel C-2 substituted vitamin D derivatives having ringed side chains and their biological evaluation on bone
J.Steroid Biochem.Mol.Biol., 136, 2013
|
|
8HTA
 
 | |
7JTW
 
 | | Crystal structure of RORgt with compound (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid | | Descriptor: | (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid, GLYCEROL, RAR-related orphan receptor C isoform a variant, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2020-08-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 6-Oxo-4-phenyl-hexanoic acid derivatives as ROR gamma t inverse agonists showing favorable ADME profile.
Bioorg.Med.Chem.Lett., 36, 2021
|
|
8GSY
 
 | |
8GSX
 
 | |
4U72
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (A260G mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
4U74
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (G262A mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
