7YJD
 
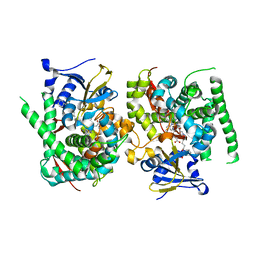 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-07-19 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
5XNE
 
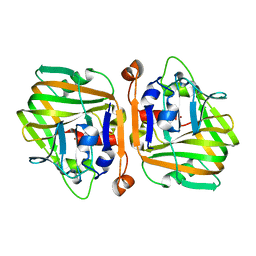 | |
4MYS
 
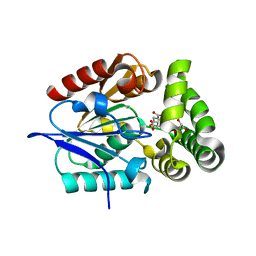 | | 1.4 Angstrom Crystal Structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase with SHCHC and Pyruvate | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, GLYCEROL, ... | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-28 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
4MYD
 
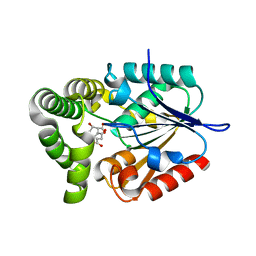 | | 1.37 Angstrom Crystal Structure of E. Coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) in complex with SHCHC | | Descriptor: | 2-(3-CARBOXYPROPIONYL)-6-HYDROXY-CYCLOHEXA-2,4-DIENE CARBOXYLIC ACID, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-27 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
4MXD
 
 | | 1.45 angstronm crystal structure of E.coli 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase (MenH) | | Descriptor: | 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sun, Y, Yin, S, Feng, Y, Li, J, Zhou, J, Liu, C, Zhu, G, Guo, Z. | | Deposit date: | 2013-09-26 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of the general base catalysis of an alpha / beta-hydrolase catalytic triad.
J.Biol.Chem., 289, 2014
|
|
8HEP
 
 | |
8HEQ
 
 | |
8HER
 
 | |
5GQQ
 
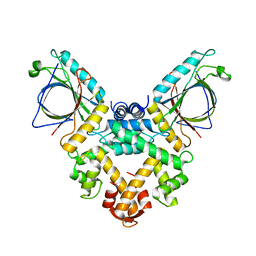 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
4I7B
 
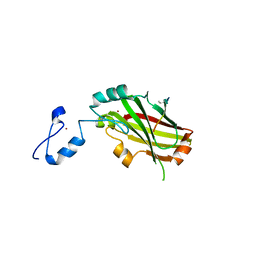 | | Siah1 bound to synthetic peptide (ACE)KLRPV(ABA)MVRPTVR | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ZINC ION | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I7C
 
 | | Siah1 mutant bound to synthetic peptide (ACE)KLRPV(23P)MVRPWVR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ... | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4I7D
 
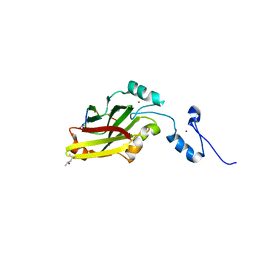 | | Siah1 bound to synthetic peptide (ACE)KLRPVAMVRP(PRK)VR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, E3 ubiquitin-protein ligase SIAH1, Protein phyllopod, ... | | Authors: | Santelli, E, Stebbins, J.L, Feng, Y, De, S.K, Purves, A, Motamedchaboki, K, Wu, B, Ronai, Z.A, Liddington, R.C, Pellecchia, M. | | Deposit date: | 2012-11-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of covalent siah inhibitors.
Chem.Biol., 20, 2013
|
|
4L4P
 
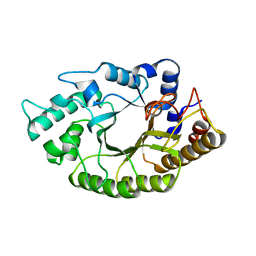 | | the mutant(E139A) structure in complex with xylotriose | | Descriptor: | Endo-1,4-beta-xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | An, J, Feng, Y, Wu, G. | | Deposit date: | 2013-06-08 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of CbXyn10B from Caldicellulosiruptor bescii and its mutant(E139A) in complex with xylotriose
To be Published
|
|
4L4O
 
 | | The crystal structure of CbXyn10B in native form | | Descriptor: | Endo-1,4-beta-xylanase, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | An, J, Feng, Y, Wu, G. | | Deposit date: | 2013-06-08 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of CbXyn10B from Caldicellulosiruptor bescii and its mutant(E139A) in complex with xylotriose
To be Published
|
|
8IGR
 
 | | Cryo-EM structure of CII-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
8IGS
 
 | | Cryo-EM structure of RNAP-promoter open complex at lambda promoter PRE | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhao, M, Gao, B, Wen, A, Feng, Y, Lu, Y. | | Deposit date: | 2023-02-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of lambda CII-dependent transcription activation.
Structure, 31, 2023
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | Descriptor: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
7XYB
 
 | | The cryo-EM structure of an AlpA-loaded complex | | Descriptor: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wen, A, Feng, Y. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7XYA
 
 | | The cryo-EM structure of an AlpA-loading complex | | Descriptor: | AlpA, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wen, A, Feng, Y. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of AlpA-dependent transcription antitermination.
Nucleic Acids Res., 50, 2022
|
|
7YWA
 
 | | Structure of DinI in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), DNA damage-inducible protein I, MAGNESIUM ION, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HOT
 
 | |
8HOO
 
 | |
8JD0
 
 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of NAM563 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCZ
 
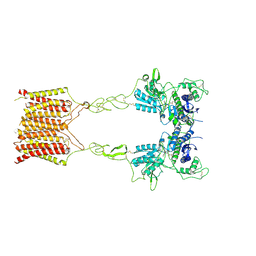 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
