9DA6
 
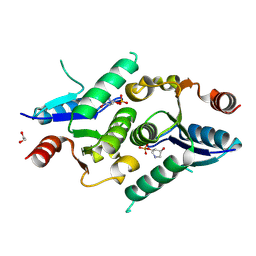 | | Crystal structure of human DNPH1 bound to inhibitor 3a | | Descriptor: | (3R,4R)-3-hydroxy-4-[(phosphonooxy)methyl]pyrrolidinium, 1,2-ETHANEDIOL, 5-hydroxymethyl-dUMP N-hydrolase | | Authors: | Wagner, A.G, Schramm, V.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-08-21 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Transition State Analogs of Human DNPH1 Reveal Two Electrophile Migration Mechanisms.
J.Med.Chem., 68, 2025
|
|
9C66
 
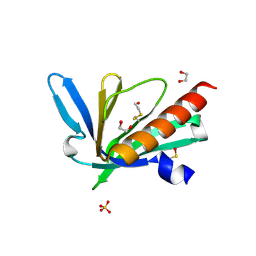 | | Structure of the Mena EVH1 domain bound to the polyproline segment of PTP1B | | Descriptor: | 1,2-ETHANEDIOL, Protein enabled homolog, SULFATE ION, ... | | Authors: | LaComb, L, Fedorov, E, Bonanno, J.B, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the Interaction Landscape of the EVH1 Domain of Mena.
Biochemistry, 63, 2024
|
|
9C6V
 
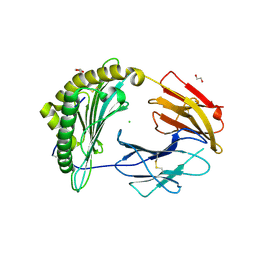 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (1 molecule/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9C6X
 
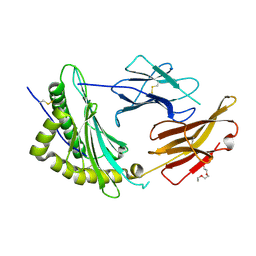 | | Crystal Structure of a single chain trimer composed of HLA-B*39:01 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, NACHT, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9C6W
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (2 molecules/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
1K0K
 
 | |
1XW5
 
 | |
1YKC
 
 | |
6C8V
 
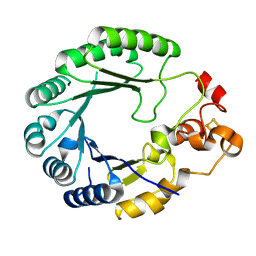 | | X-ray structure of PqqE from Methylobacterium extorquens | | Descriptor: | Coenzyme PQQ synthesis protein E, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Gizzi, A.S, Grove, T.L, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray and EPR Characterization of the Auxiliary Fe-S Clusters in the Radical SAM Enzyme PqqE.
Biochemistry, 57, 2018
|
|
6DXP
 
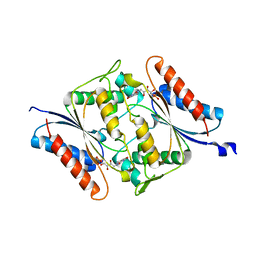 | | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Arcinas, A.J, Ghosh, A, Chamala, S, Bonanno, J.B, Kelly, L, Almo, S.C. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | The crystal structure of an FMN-dependent NADH-azoreductase from Klebsiella pneumoniae
To Be Published
|
|
6CIA
 
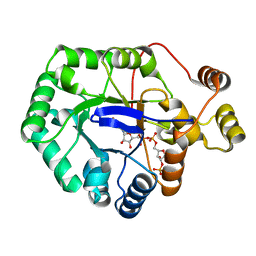 | | Crystal structure of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH. | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lipowska, J, Leung, E.S, Shabalin, I.G, Grabowski, M, Almo, S.C, Satchell, K.J, Joachimiak, A, Lewinski, K, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-23 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of of aldo-keto reductase from Klebsiella pneumoniae in complex with NADPH.
to be published
|
|
1XW6
 
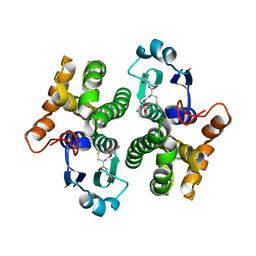 | | 1.9 angstrom resolution structure of human glutathione S-transferase M1A-1A complexed with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-10-29 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
1XWK
 
 | | 2.3 angstrom resolution crystal structure of human glutathione S-transferase M1A-1A complexed with glutathionyl-S-dinitrobenzene | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-11-01 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
1YEY
 
 | | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | L-fuconate dehydratase, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-28 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of L-fuconate Dehydratase from Xanthomonas campestris pv. campestris str. ATCC 33913
To be Published
|
|
1YJ6
 
 | |
4YZZ
 
 | | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a TRAP transporter solute binding protein (IPR025997) from Bordetella bronchiseptica RB50 (BB0280, TARGET EFI-500035) mixed occupancy dimer, copurified calcium and picolinate bound active site versus apo site
To be published
|
|
4ZJP
 
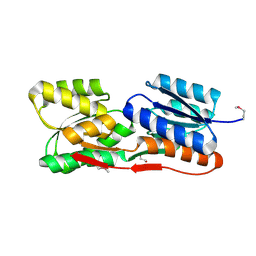 | | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose | | Descriptor: | 1,2-ETHANEDIOL, Monosaccharide-transporting ATPase, beta-D-ribopyranose | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Glenn, A.S, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of an ABC-Transporter Solute Binding Protein (SBP_IPR025997) from Actinobacillus Succinogenes (Asuc_0197, TARGET EFI-511067) with bound beta-D-ribopyranose
To be published
|
|
4ZQB
 
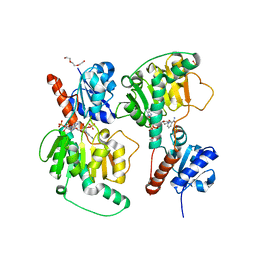 | | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kowiel, M, Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Porebski, P.J, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-08 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of NADP-dependent dehydrogenase from Rhodobactersphaeroides in complex with NADP and sulfate
to be published
|
|
5VE2
 
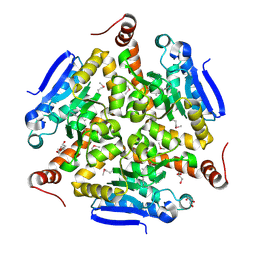 | | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase, GLYCEROL, ... | | Authors: | Siuda, M.K, Shabalin, I.G, Cooper, D.R, Chapman, H.C, Tkaczuk, K.L, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of enoyl-CoA hydratase/isomerase from Pseudoalteromonas atlantica T6c at 2.3 A resolution.
to be published
|
|
5BP7
 
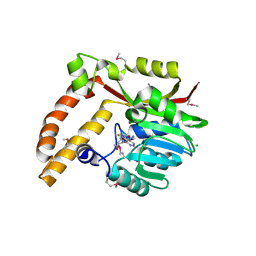 | | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kutner, J, Shabalin, I.G, Mason, D.V, Handing, K.B, Gasiorowska, O.A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of SAM-dependent methyltransferase from Geobacter sulfurreducens in complex with S-Adenosyl-L-homocysteine
to be published
|
|
4Z18
 
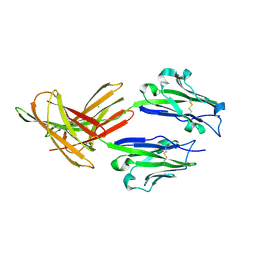 | | CRYSTAL STRUCTURE OF HUMAN PD-L1 | | Descriptor: | CHLORIDE ION, Programmed cell death 1 ligand 1 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samantha, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | CRYSTAL STRUCTURE OF HUMAN PD-L1
To Be Published
|
|
4ZQ8
 
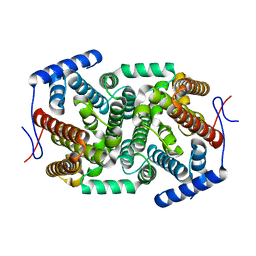 | | Crystal structure of a terpene synthase from Streptomyces lydicus, target EFI-540129 | | Descriptor: | Isoprenoid Synthase | | Authors: | Toro, R, Bhosle, R, Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Poulter, C.D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-05-08 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a terpene synthase from Streptomyces lydicus, target EFI-540129
To be published
|
|
4Z0N
 
 | | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a Periplasmic Solute binding protein (IPR025997) from Streptobacillus moniliformis DSM-12112 (Smon_0317, TARGET EFI-511281) with bound D-Galactose
To be published
|
|
7MJV
 
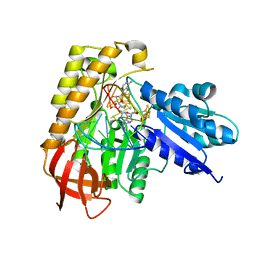 | | MiaB in the complex with s-adenosylmethionine and RNA | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
7MJY
 
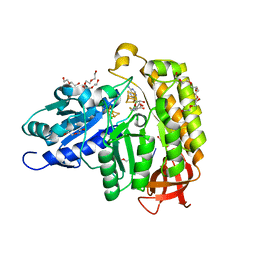 | | MiaB in the complex with s-adenosyl-L-homocysteine and RNA | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, ... | | Authors: | Esakova, O.A, Grove, T.L, Yennawar, N.H, Arcinas, A.J, Wang, B, Krebs, C, Almo, S.C, Booker, S.J. | | Deposit date: | 2021-04-20 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB.
Nature, 597, 2021
|
|
