5Z0Y
 
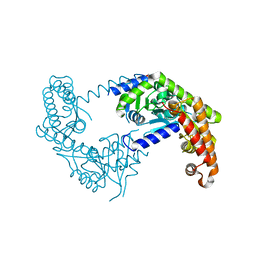 | |
6A57
 
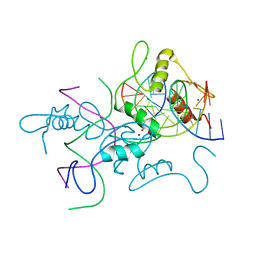 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
5ZBN
 
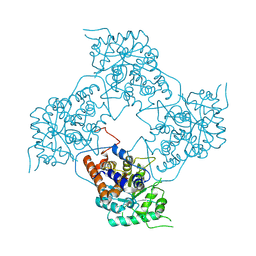 | |
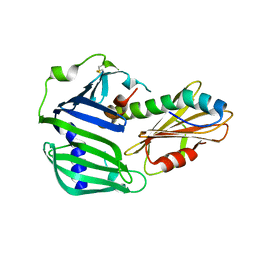
6IN8
 
 | | Crystal structure of MucB | | Descriptor: | Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6IN9
 
 | | Crystal structure of MucB in complex with MucA(peri) | | Descriptor: | Sigma factor AlgU negative regulatory protein, Sigma factor AlgU regulatory protein MucB | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6IN7
 
 | | Crystal structure of AlgU in complex with MucA(cyto) | | Descriptor: | NICOTINAMIDE, RNA polymerase sigma-H factor, Sigma factor AlgU negative regulatory protein | | Authors: | Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for the recognition of MucA by MucB and AlgU in Pseudomonas aeruginosa.
Febs J., 286, 2019
|
|
6JDS
 
 | |
6JDU
 
 | | Crystal structure of PRRSV nsp10 (helicase) | | Descriptor: | CALCIUM ION, PP1b, ZINC ION | | Authors: | Tang, C, Chen, Z. | | Deposit date: | 2019-02-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Helicase of Type 2 Porcine Reproductive and Respiratory Syndrome Virus Strain HV Reveals a Unique Structure.
Viruses, 12, 2020
|
|
6JDR
 
 | |
6KXS
 
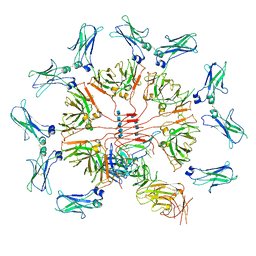 | | Cryo-EM structure of human IgM-Fc in complex with the J chain and the ectodomain of pIgR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Li, Y, Wang, G, Xiao, J. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into immunoglobulin M.
Science, 367, 2020
|
|
8Z3K
 
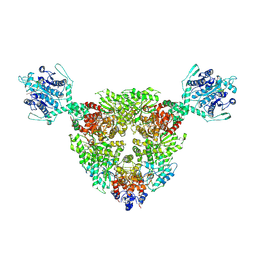 | | The structure of type III CRISPR-associated deaminase in complex 2cA6-2ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-11 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
8Z40
 
 | |
8Z3R
 
 | | The structure of type III CRISPR-associated deaminase in complex cA4 | | Descriptor: | 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Adenosine deaminase domain-containing protein, ZINC ION | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-16 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
8Z3P
 
 | | The structure of type III CRISPR-associated deaminase in complex cA6 and ATP, fully activated | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenosine deaminase domain-containing protein, MAGNESIUM ION, ... | | Authors: | Chen, M.R, Li, Z.X, Xiao, Y.B. | | Deposit date: | 2024-04-15 | | Release date: | 2024-12-25 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Antiviral signaling of a type III CRISPR-associated deaminase.
Science, 387, 2025
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
7VJV
 
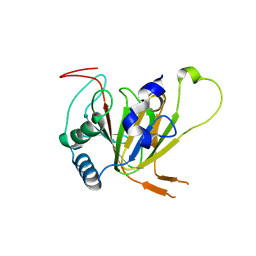 | | Human AlkB homolog ALKBH6 in complex with alpha-katoglutarate and Mn | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, MANGANESE (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJS
 
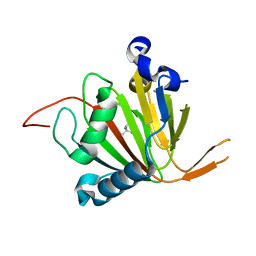 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7V6X
 
 | | Crystal structure of HPPD complexed with Y18556 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-3-[(2-methylphenyl)methyl]-6-[(1~{R},2~{S})-2-oxidanyl-6-oxidanylidene-cyclohexyl]carbonyl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Lin, H.Y, Yang, G.F. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pharmacophore-Oriented Discovery of Novel 1,2,3-Benzotriazine-4-one Derivatives as Potent 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 70, 2022
|
|
7VW9
 
 | |
7TZ6
 
 | | Structure of mitochondrial bc1 in complex with ck-2-68 | | Descriptor: | 7-chloranyl-3-methyl-2-[4-[[4-(trifluoromethyloxy)phenyl]methyl]phenyl]-1~{H}-quinolin-4-one, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Huang, R. | | Deposit date: | 2022-02-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of complex III with bound antimalarial agent CK-2-68 provides insights into selective inhibition of Plasmodium cytochrome bc 1 complexes.
J.Biol.Chem., 299, 2023
|
|
7VWA
 
 | |
7VW8
 
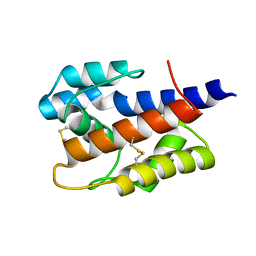 | |
7WPQ
 
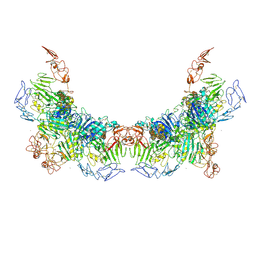 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 3.27 angstron resolution (2 units) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.267 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WPP
 
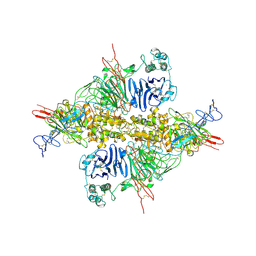 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 2.85 angstron resolution (1 unit) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
7WQT
 
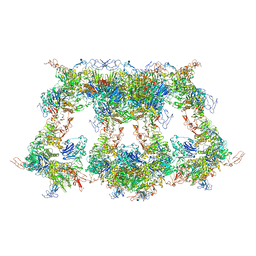 | | Cryo-EM structure of VWF D'D3 dimer complexed with D1D2 at 4.3 angstron resolution (VWF tube) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand antigen 2, ... | | Authors: | Zeng, J.W, Shu, Z.M, Zhou, A.W. | | Deposit date: | 2022-01-26 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of von Willebrand factor multimerization and tubular storage.
Blood, 139, 2022
|
|
