8A6F
 
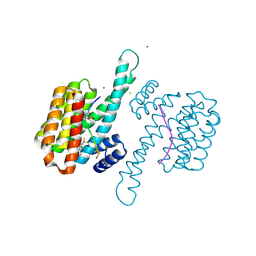 | | Small molecule stabilizer (compound 8) for C-RAF and 14-3-3 | | Descriptor: | 1-[4-[4-chloranyl-3-(trifluoromethyl)phenyl]-4-oxidanyl-piperidin-1-yl]-3-[2-(dimethylamino)ethyldisulfanyl]propan-1-one, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Kenanova, D.N, Visser, E.J, Virta, J, Sijbesma, E, Centorrino, F, Zhong, M, Vickery, H, Neitz, J, Brunsveld, L, Ottmann, C, Arkin, M.R. | | Deposit date: | 2022-06-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Acs Cent.Sci., 9, 2023
|
|
6ZI3
 
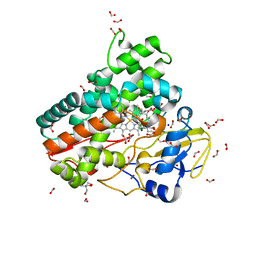 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | Authors: | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | Deposit date: | 2020-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
8COI
 
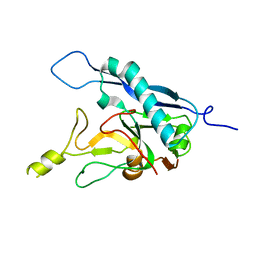 | | Human adenovirus-derived synthetic ADDobody binder | | Descriptor: | ADDobody | | Authors: | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8AAJ
 
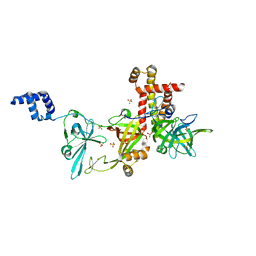 | | Crystal structure of the Pyrococcus abyssi RPA (apo form) | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Legrand, P, Madru, C, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8AAS
 
 | | Crystal structure of the Pyrococcus abyssi RPA trimerization core bound to poly-dT20 ssDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, ... | | Authors: | Madru, C, Legrand, P, Sauguet, L. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8AA9
 
 | |
8ACT
 
 | | structure of the human beta-cardiac myosin folded-back off state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin light chain 3, ... | | Authors: | Grinzato, A, Kandiah, E, Robert-Paganin, J, Auguin, D, Kikuti, C, Nandwani, N, Moussaoui, D, Pathak, D, Ruppel, K.M, Spudich, J.A, Houdusse, A. | | Deposit date: | 2022-07-06 | | Release date: | 2023-06-07 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the folded-back state of human beta-cardiac myosin.
Nat Commun, 14, 2023
|
|
6F27
 
 | | NMR solution structure of non-bound [des-Arg10]-kallidin (DAKD) | | Descriptor: | DAKD | | Authors: | Richter, C, Jonker, H.R.A, Schwalbe, H, Joedicke, L, Mao, J, Kuenze, G, Reinhart, C, Kalavacherla, T, Meiler, J, Preu, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
8A1G
 
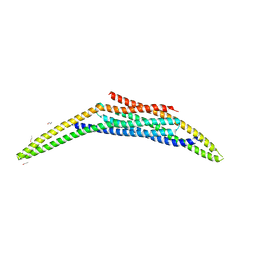 | | Structure of the SNX1-SNX5 complex | | Descriptor: | N-PROPANOL, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E.N, Banos-Mateos, S, Vidaurrazaga, A, Rojas, A.L, Castano, D, Hierro, A. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A11
 
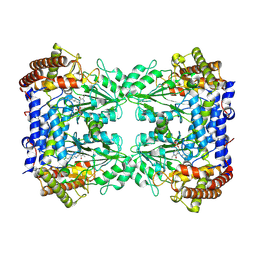 | | Cryo-EM structure of the Human SHMT1-RNA complex | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, cytosolic | | Authors: | Spizzichino, S, Marabelli, C, Bharadwaj, A, Jakobi, A.J, Chaves-Sanjuan, A, Giardina, G, Bolognesi, M, Cutruzzola, F. | | Deposit date: | 2022-05-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structure-based mechanism of riboregulation of the metabolic enzyme SHMT1.
Mol.Cell, 2024
|
|
8A12
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
8ABQ
 
 | | Structure of the SNX1-SNX5 complex, Pt derivative | | Descriptor: | PLATINUM (II) ION, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E.N, Banos-Mateos, S, Vidaurrazaga, A, Rojas, A.L, Castano, D, Hierro, A. | | Deposit date: | 2022-07-04 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AFZ
 
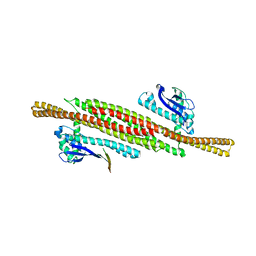 | | Architecture of the ESCPE-1 membrane coat | | Descriptor: | Cation-independent mannose-6-phosphate receptor, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E, Ishida, M, Williamsom, C.D, Banos-Mateos, S, Gil-Carton, D, Romero, M, Vidaurrazaga, A, Fernandez-Recio, J, Rojas, A.L, Bonifacino, J.S, Castano-Diez, D, Hierro, A. | | Deposit date: | 2022-07-18 | | Release date: | 2023-06-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1CUV
 
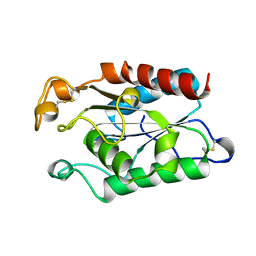 | | CUTINASE, A85F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
8AIL
 
 | | Bacillus phage VMY22 p56 in complex with Bacillus weidmannii Ung | | Descriptor: | Bacillus phage VMY22 p56, GLYCEROL, IODIDE ION, ... | | Authors: | Muselmani, W, Bagneris, C, Savva, R. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
1CUJ
 
 | | CUTINASE, S120C MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
8AN4
 
 | | MenT1 toxin (rv0078a) from Mycobacterium tuberculosis H37Rv | | Descriptor: | Bacterial toxin | | Authors: | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
8B78
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{R})-8-(2-chloranyl-6-oxidanyl-phenyl)-7-fluoranyl-9-prop-1-ynyl-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
8AN5
 
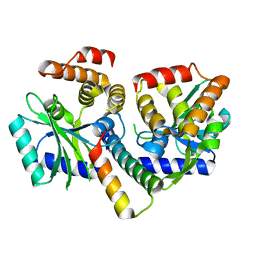 | | MenAT1 toxin-antitoxin complex (rv0078a-rv0078b) from Mycobacterium tuberculosis H37Rv | | Descriptor: | Bacterial toxin, Conserved protein | | Authors: | Xu, X, Usher, B, Gutierrez, C, Barriot, R, Arrowsmith, T.J, Han, X, Redder, P, Neyrolles, O, Blower, T.R, Genevaux, P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | MenT nucleotidyltransferase toxins extend tRNA acceptor stems and can be inhibited by asymmetrical antitoxin binding.
Nat Commun, 14, 2023
|
|
8B6I
 
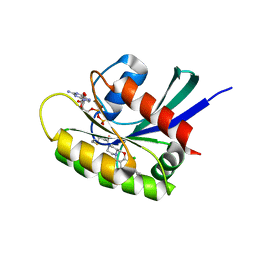 | | KRasG12C ligand complex | | Descriptor: | 1-[(4~{a}~{S})-7-chloranyl-8-(5-methyl-2~{H}-indazol-4-yl)-1,2,4,4~{a},5,11-hexahydropyrazino[2,1-c][1,4]benzoxazepin-3-yl]propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of AZD4747, a Potent and Selective Inhibitor of Mutant GTPase KRAS G12C with Demonstrable CNS Penetration.
J.Med.Chem., 66, 2023
|
|
6YWF
 
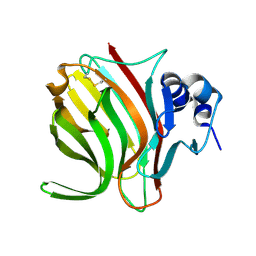 | |
2OPG
 
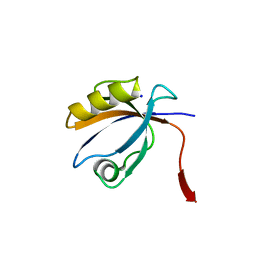 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
8AK5
 
 | |
8AKB
 
 | |
8AKF
 
 | |
