6UOE
 
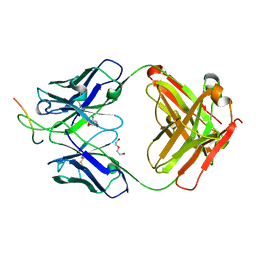 | |
6VC9
 
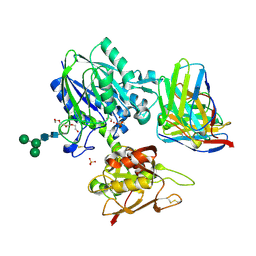 | | TB19 complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-nucleotidase, ecto (CD73), ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
6VCA
 
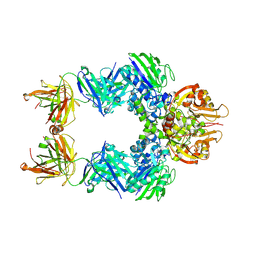 | | TB38 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, ... | | Authors: | Zhou, Y.F, Lord, D.M. | | Deposit date: | 2019-12-20 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | A highly potent CD73 biparatopic antibody blocks organization of the enzyme active site through dual mechanisms.
J.Biol.Chem., 295, 2020
|
|
6WYV
 
 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
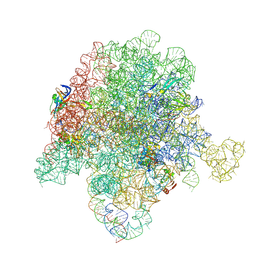
6X3C
 
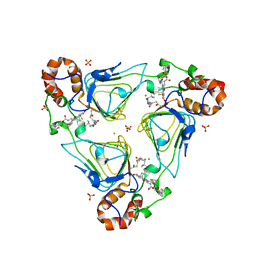 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F1037 (47) | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
2I0N
 
 | | Structure of Dictyostelium discoideum Myosin VII SH3 domain with adjacent proline rich region | | Descriptor: | Class VII unconventional myosin | | Authors: | Wang, Q, Deloia, M.A, Kang, Y, Litchke, C, Titus, M.A, Walters, K.J. | | Deposit date: | 2006-08-10 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The SH3 domain of a M7 interacts with its C-terminal proline-rich region.
Protein Sci., 16, 2007
|
|
2KBJ
 
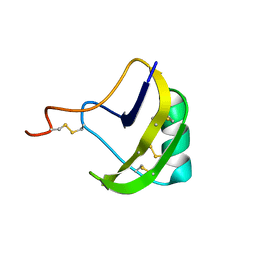 | | solution structure of BmKalphaTx11 (minor conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
6M3G
 
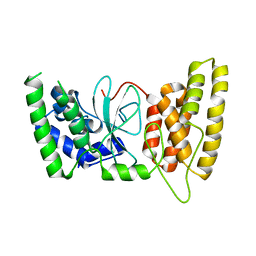 | | Crystal structure of human HPF1 | | Descriptor: | Histone PARylation factor 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6M3I
 
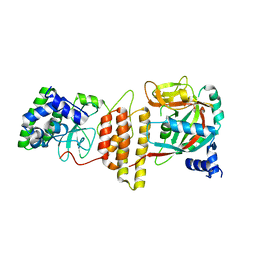 | | Crystal structure of HPF1/PARP1 complex | | Descriptor: | BENZAMIDE, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6M3H
 
 | | Crystal structure of mouse HPF1 | | Descriptor: | Histone PARylation factor 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6LUO
 
 | | Structure of nurse shark beta-2-microglobulin | | Descriptor: | Beta-2-microglobulin | | Authors: | Xia, C, Wu, Y. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6LUP
 
 | | Crystal structure of shark MHC CLASS I for 2.3 angstrom | | Descriptor: | Beta-2-microglobulin, MHC class I protein, PHE-ALA-ASN-PHE-PHE-ILE-ARG-GLY-LEU | | Authors: | Wu, Y, Xia, C. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6N2G
 
 | | Crystal structure of Caenorhabditis elegans NAP1 | | Descriptor: | Nucleosome Assembly Protein | | Authors: | Bhattacharyya, S, DArcy, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Characterization of Caenorhabditis elegans Nucleosome Assembly Protein 1 Uncovers the Role of Acidic Tails in Histone Binding.
Biochemistry, 58, 2019
|
|
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E7X
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7CQW
 
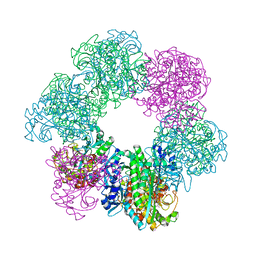 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQQ
 
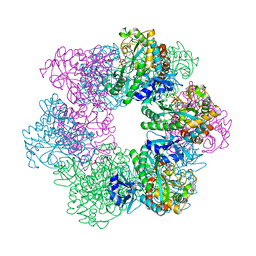 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQL
 
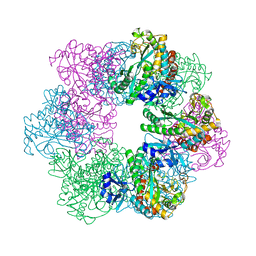 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
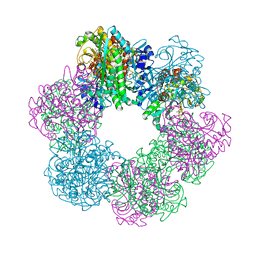 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
2KBH
 
 | | solution structure of BmKalphaTx11 (major conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2N3T
 
 | |
2N3W
 
 | |
