4M7P
 
 | | Ensemble refinement of protein crystal structure of macrolide glycosyltransferases OleD | | Descriptor: | Oleandomycin glycosyltransferase, SODIUM ION | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
4K3D
 
 | | Crystal structure of bovine antibody BLV1H12 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K3E
 
 | | Crystal structure of bovine antibody BLV5B8 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
4WS9
 
 | | Crystal structure of sMAT N159G from Sulfolobus solfataricus | | Descriptor: | PHOSPHATE ION, S-adenosylmethionine synthase | | Authors: | Wang, F, Brady, E.L, Singh, S, Clinger, J.A, Huber, T.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2014-10-26 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of sMAT N159G from Sulfolobus solfataricus.
To Be Published
|
|
5CET
 
 | | Crystal structure of Rv2837c | | Descriptor: | Bifunctional oligoribonuclease and PAP phosphatase NrnA, MANGANESE (II) ION | | Authors: | Wang, F, He, Q, Zhu, D, Liu, S, Gu, L. | | Deposit date: | 2015-07-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Insight into the Mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP Phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
5GT9
 
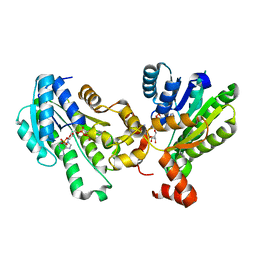 | | The X-ray structure of 7beta-hydroxysteroid dehydrogenase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Wang, F, Wang, R, Lv, Z, Chen, Q, Huo, X. | | Deposit date: | 2016-08-19 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of NADP(+)-bound 7 beta-hydroxysteroid dehydrogenase reveals two cofactor-binding modes
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6MK1
 
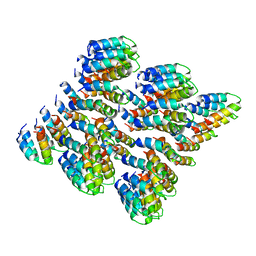 | | Cryo-EM of self-assembly peptide filament HEAT_R1 | | Descriptor: | peptide HEAT_R1 | | Authors: | Wang, F, Hughes, S.A, Orlova, A, Conticello, V.P, Egelman, E.H. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Ambidextrous helical nanotubes from self-assembly of designed helical hairpin motifs.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2GIZ
 
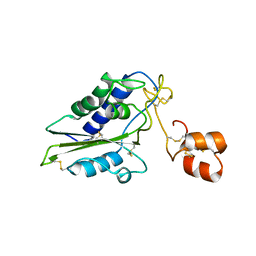 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
8FOF
 
 | |
7T6E
 
 | |
7TXI
 
 | | Cryo-EM of A. pernix flagellum | | Descriptor: | Probable flagellin 1 | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UUQ
 
 | |
3FNH
 
 | | Crystal structure of InhA bound to triclosan derivative | | Descriptor: | 2-(2,4-DICHLOROPHENOXY)-5-(2-PHENYLETHYL)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F. | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Triclosan Derivatives: Towards Potent Inhibitors of Drug-Sensitive and Drug-Resistant Mycobacterium tuberculosis.
Chemmedchem, 4, 2009
|
|
3FNE
 
 | | Crystal structure of InhA bound to triclosan derivative 17 | | Descriptor: | 2-(2,4-DICHLOROPHENOXY)-5-(PYRIDIN-2-YLMETHYL)PHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F. | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Triclosan Derivatives: Towards Potent Inhibitors of Drug-Sensitive and Drug-Resistant Mycobacterium tuberculosis.
Chemmedchem, 4, 2009
|
|
3FCA
 
 | | Genetic Incorporation of a Metal-ion Chelating Amino Acid into proteins as biophysical probe | | Descriptor: | Cysteine synthase, ZINC ION | | Authors: | Wang, F, Lee, H, Spraggon, G, Schultz, P.G. | | Deposit date: | 2008-11-21 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe.
J.Am.Chem.Soc., 131, 2009
|
|
8FK7
 
 | | Structure of the Pyrobaculum calidifontis flagellar-like archaeal type IV pilus | | Descriptor: | Flagellin | | Authors: | Wang, F, Kreutzberger, M.A, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FJ5
 
 | | Structure of the Haloferax volcanii archaeal type IV pilus | | Descriptor: | Pilin_N domain-containing protein | | Authors: | Wang, F, Kreutzberger, M.A, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3FNF
 
 | | Crystal structure of InhA bound to triclosan derivative | | Descriptor: | 5-benzyl-2-(2,4-dichlorophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F. | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Triclosan Derivatives: Towards Potent Inhibitors of Drug-Sensitive and Drug-Resistant Mycobacterium tuberculosis.
Chemmedchem, 4, 2009
|
|
6UHZ
 
 | | WDR5 in complex with Myc site fragment inhibitor | | Descriptor: | 1-cyclohexyl-1H-benzotriazole-5-carboxylic acid, WDR5 | | Authors: | Wang, F, Fesik, S.w. | | Deposit date: | 2019-09-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | Discovery of WD Repeat-Containing Protein 5 (WDR5)-MYC Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6UHY
 
 | | WDR5 in complex with Myc site fragment inhibitor | | Descriptor: | 1-cyclohexyl-1H-benzimidazole-5-carboxylic acid, WDR5 | | Authors: | Wang, F, Fesik, S.W. | | Deposit date: | 2019-09-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of WD Repeat-Containing Protein 5 (WDR5)-MYC Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
7DLV
 
 | | shrimp dUTPase in complex with Stl | | Descriptor: | CALCIUM ION, Orf20, SULFATE ION, ... | | Authors: | Ma, Q, Wang, F. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Structural basis of staphylococcal Stl inhibition on a eukaryotic dUTPase.
Int.J.Biol.Macromol., 184, 2021
|
|
8EWG
 
 | | Cryo-EM structure of a riboendonclease | | Descriptor: | CRISPR-associated endonuclease Cas9, RNA (56-MER) | | Authors: | Li, Z, Wang, F. | | Deposit date: | 2022-10-23 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural Basis for the Ribonuclease Activity of a Thermostable CRISPR-Cas13a from Thermoclostridium caenicola.
J.Mol.Biol., 435, 2023
|
|
6VY1
 
 | | Cryo-EM structure of filamentous PFD from Methanocaldococcus jannaschii | | Descriptor: | Prefoldin subunit alpha 2 | | Authors: | Wang, F, Chen, Y.X, Ing, N.L, Hochbaum, A.I, Clark, D.S, Glover, D.J, Egelman, E.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Determination of a Filamentous Chaperone to Fabricate Electronically Conductive Metalloprotein Nanowires.
Acs Nano, 14, 2020
|
|
3FNG
 
 | | Crystal structure of InhA bound to triclosan derivative | | Descriptor: | 5-(cyclohexylmethyl)-2-(2,4-dichlorophenoxy)phenol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, F. | | Deposit date: | 2008-12-24 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Triclosan Derivatives: Towards Potent Inhibitors of Drug-Sensitive and Drug-Resistant Mycobacterium tuberculosis.
Chemmedchem, 4, 2009
|
|
5D6Y
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
