4KPX
 
 | | Hin GlmU bound to WG766 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, N-{4-[(4-hydroxy-3-nitrobenzoyl)amino]phenyl}pyridine-2-carboxamide, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-14 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
6JSJ
 
 | |
4KNX
 
 | | Hin GlmU Bound to WG176 | | Descriptor: | Bifunctional protein GlmU, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Doig, P, Kazmirski, S.L, Boriack-Sjodin, P.A. | | Deposit date: | 2013-05-10 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of inhibitors of the bacterial cell wall synthetic enzyme GlmU using virtual screening and lead-hopping.
Bioorg.Med.Chem., 22, 2014
|
|
5GNU
 
 | | the structure of mini-MFN1 apo | | Descriptor: | Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Lou, J. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.113 Å) | | Cite: | BDLP-like folding of Mitofusin 1
To Be Published
|
|
6ADM
 
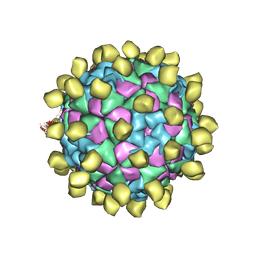 | |
7E2U
 
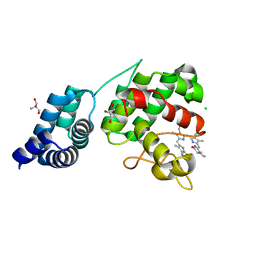 | | Synechocystis GUN4 in complex with phytochrome | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2T
 
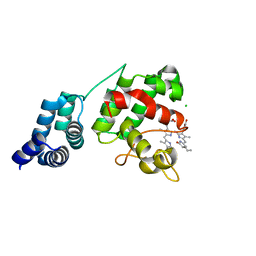 | | Synechocystis GUN4 in complex with phycocyanobilin | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2S
 
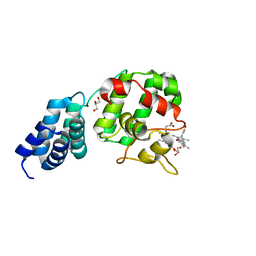 | | Synechocystis GUN4 in complex with biliverdin IXa | | Descriptor: | BILIVERDINE IX ALPHA, GLYCEROL, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
6ADR
 
 | |
6ADL
 
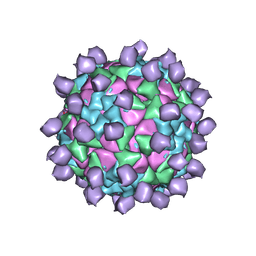 | |
6U2D
 
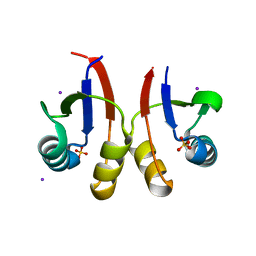 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
6ADS
 
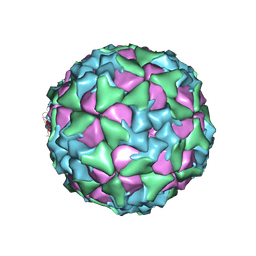 | | Structure of Seneca Valley Virus in acidic conditions | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ADT
 
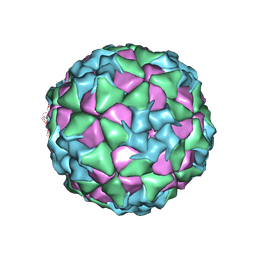 | | Structure of Seneca Valley Virus in neutral condition | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Lou, Z.Y, Cao, L. | | Deposit date: | 2018-08-02 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Seneca Valley virus attachment and uncoating mediated by its receptor anthrax toxin receptor 1
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4XWW
 
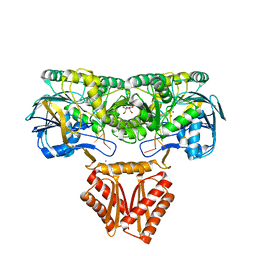 | | Crystal structure of RNase J complexed with RNA | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
6JFK
 
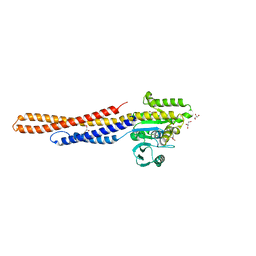 | | GDP bound Mitofusin2 (MFN2) | | Descriptor: | CITRIC ACID, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
6JFL
 
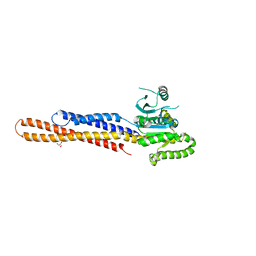 | | Nucleotide-free Mitofusin2 (MFN2) | | Descriptor: | CALCIUM ION, GLYCEROL, Mitofusin-2,cDNA FLJ57997, ... | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
4XWT
 
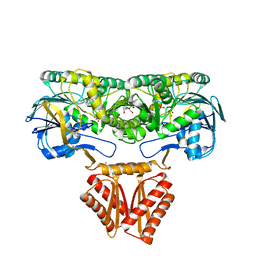 | | Crystal structure of RNase J complexed with UMP | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
6JFM
 
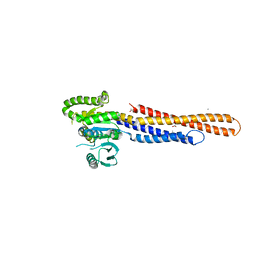 | | Mitofusin2 (MFN2)_T111D | | Descriptor: | ACETATE ION, CALCIUM ION, Mitofusin-2,Mitofusin-2 | | Authors: | Li, Y.J, Cao, Y.L, Feng, J.X, Qi, Y.B, Meng, S.X, Yang, J.F, Zhong, Y.T, Kang, S.S, Chen, X.X, Lan, L, Luo, L, Yu, B, Chen, S.D, Chan, D.C, Hu, J.J, Gao, S. | | Deposit date: | 2019-02-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural insights of human mitofusin-2 into mitochondrial fusion and CMT2A onset.
Nat Commun, 10, 2019
|
|
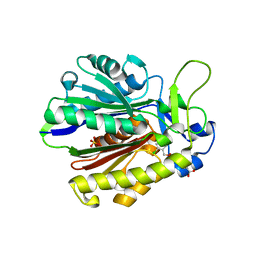
4Z8D
 
 | |
5BNM
 
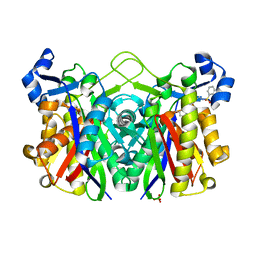 | | E. coli FabH with Small Molecule Inhibitor 1 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, N-{[3'-(hydroxymethyl)biphenyl-4-yl]methyl}benzenesulfonamide, SULFATE ION, ... | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5BNR
 
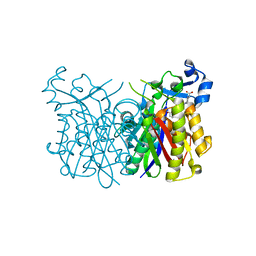 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5WSH
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, GLY-VAL-TRP-ILE-ARG-THR-PRO-THR-ALA, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2016-12-07 | | Release date: | 2017-12-20 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
5BQS
 
 | | S. Pneumoniae Fabh with small molecule inhibitor 4 | | Descriptor: | 1-{5-[2-chloro-5-(hydroxymethyl)phenyl]pyridin-2-yl}piperidine-4-carboxylic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 3, SODIUM ION | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-29 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
5BNS
 
 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}-N-(quinolin-6-ylmethyl)piperidine-4-carboxamide, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
8J6S
 
 | | Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
