4BXD
 
 | |
4BXJ
 
 | |
4BPA
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with NAG-NAM-NAG-NAM tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, AMPDH2, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E, Lastochkin, E, Zhang, W, Hellman, L, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4CAJ
 
 | | Crystallographic structure of the mouse SIGN-R1 CRD domain in complex with sialic acid | | Descriptor: | CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, CHLORIDE ION, ... | | Authors: | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2013-10-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
4BXE
 
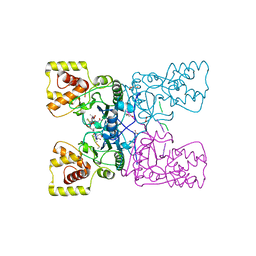 | |
4CFP
 
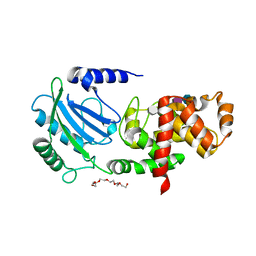 | | Crystal structure of MltC in complex with tetrasaccharide at 2.15 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-beta-muramic acid-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-methyl 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-beta-D-glucopyranoside, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Bernardo-Garcia, N, Artola-Recolons, C, Hermoso, J.A. | | Deposit date: | 2013-11-19 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4BUR
 
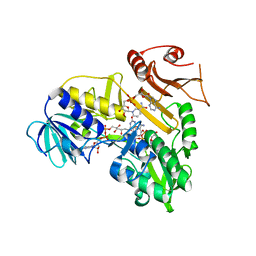 | | Crystal structure of the reduced human Apoptosis inducing factor complexed with NAD | | Descriptor: | APOPTOSIS INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4C9F
 
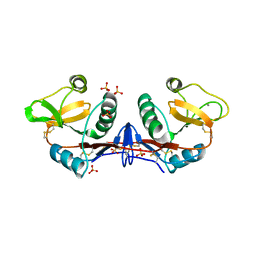 | | Structure of SIGN-R1 in complex with Sulfodextran | | Descriptor: | 4-O-sulfo-alpha-D-glucopyranose, CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, ... | | Authors: | Silva-Martin, N, Bartual, S.G, Rodriguez, A, Ramirez, E, Chacon, P, Anthony, R.M, Park, C.G, Hermoso, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
4BV6
 
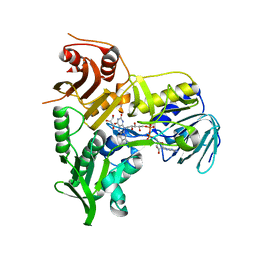 | | Refined crystal structure of the human Apoptosis inducing factor | | Descriptor: | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4C5F
 
 | |
4CHX
 
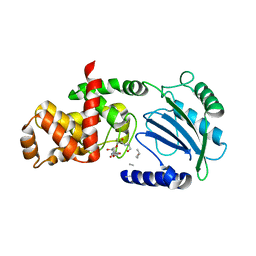 | | Crystal structure of MltC in complex with disaccharide pentapeptide DHl89 | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase C, ... | | Authors: | Artola-Recolons, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-12-04 | | Release date: | 2014-07-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Cell Wall Cleavage by Modular Lytic Transglycosylase Mltc of Escherichia Coli.
Acs Chem.Biol., 9, 2014
|
|
4CGK
 
 | | Crystal structure of the essential protein PcsB from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Bartual, S.G, Straume, D, Stamsas, G.A, Alfonso, C, Martinez-Ripoll, M, Havarstein, L.S, Hermoso, J.A. | | Deposit date: | 2013-11-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Pcsb-Mediated Cell Separation in Streptococcus Pneumoniae.
Nat.Commun., 5, 2014
|
|
4CNL
 
 | | Crystal structure of the Choline-binding domain of CbpL from Streptococcus pneumoniae | | Descriptor: | CHOLINE ION, GLYCEROL, PUTATIVE PNEUMOCOCCAL SURFACE PROTEIN, ... | | Authors: | Gutierrez-Fernandez, J, Bartual, S.G, Hermoso, J.A. | | Deposit date: | 2014-01-23 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Modular Architecture and Unique Teichoic Acid Recognition Features of Choline-Binding Protein L (Cbpl) Contributing to Pneumococcal Pathogenesis.
Sci.Rep., 6, 2016
|
|
4CPK
 
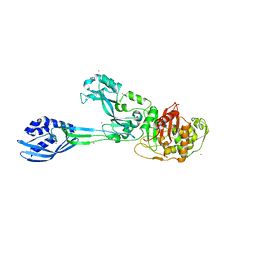 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
1BJK
 
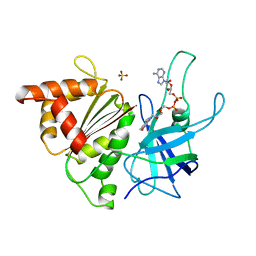 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
1BQE
 
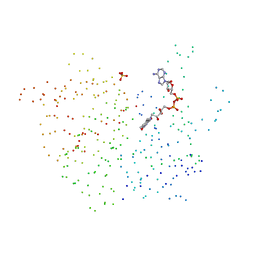 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH THR 155 REPLACED BY GLY (T155G) | | Descriptor: | FERREDOXIN--NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-08-14 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Probing the determinants of coenzyme specificity in ferredoxin-NADP+ reductase by site-directed mutagenesis.
J.Biol.Chem., 276, 2001
|
|
1GJR
 
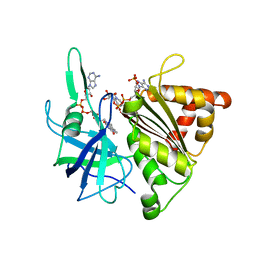 | | Ferredoxin-NADP+ Reductase complexed with NADP+ by COCRYSTALLIZATION | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2001-08-01 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Coenzyme Recognition and Binding Revealed by Crystal Structure Analysis of Ferredoxin-Nadp(+) Reductase Complexed with Nadp(+)
J.Mol.Biol., 319, 2002
|
|
8QM0
 
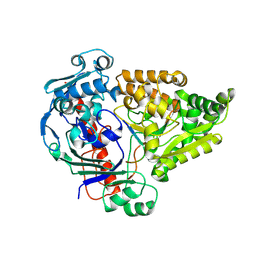 | | Crystal structure of the pneumococcal Substrate-binding protein AmiA in complex with Peptide 5 | | Descriptor: | ALA-LYS-THR-ILE-LYS-ILE-THR-GLN-THR-ARG, Oligopeptide-binding protein AmiA | | Authors: | Alcorlo, M, Abdullah, M.R, Hammerschmidt, S, Hermoso, J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular and structural basis of oligopeptide recognition by the Ami transporter system in pneumococci.
Plos Pathog., 20, 2024
|
|
8C9J
 
 | | Crystal structure of human NQO1 by serial femtosecond crystallography | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Pey, A, Botha, S, Ros, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
3ESZ
 
 | | K2AK3A Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
8I6O
 
 | |
8I6R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Xu, X, Li, J, Luo, M. | | Deposit date: | 2023-01-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanistic insights into the regulation of cell wall hydrolysis by FtsEX and EnvC at the bacterial division site.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I6S
 
 | | Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Xu, X, Li, J, Luo, M. | | Deposit date: | 2023-01-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanistic insights into the regulation of cell wall hydrolysis by FtsEX and EnvC at the bacterial division site.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I6Q
 
 | |
8PW2
 
 | |
