5W5Y
 
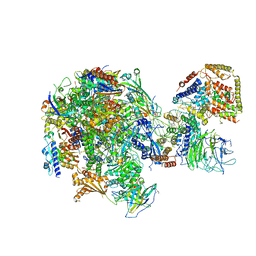 | | RNA polymerase I Initial Transcribing Complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W65
 
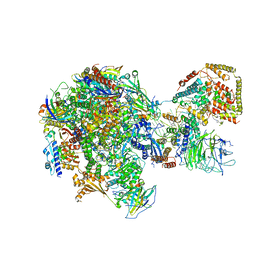 | | RNA polymerase I Initial Transcribing Complex State 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W64
 
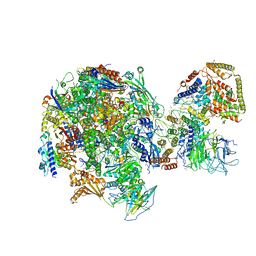 | | RNA Polymerase I Initial Transcribing Complex State 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
8EZB
 
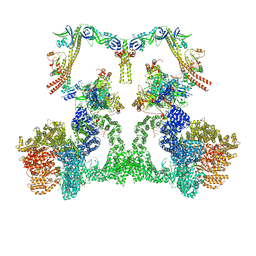 | | NHEJ Long-range complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZA
 
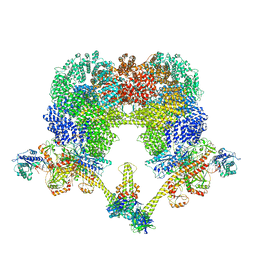 | | NHEJ Long-range complex with PAXX | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (30-MER), DNA (31-MER), ... | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
8EZ9
 
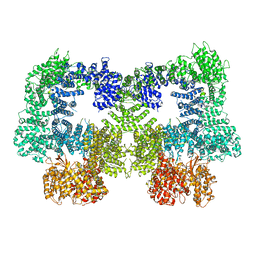 | | Dimeric complex of DNA-PKcs | | Descriptor: | DNA-dependent protein kinase catalytic subunit, unknown region of DNA-PKcs | | Authors: | Chen, S, He, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (5.67 Å) | | Cite: | Cryo-EM visualization of DNA-PKcs structural intermediates in NHEJ.
Sci Adv, 9, 2023
|
|
3L8J
 
 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
3L8I
 
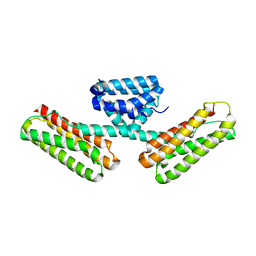 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
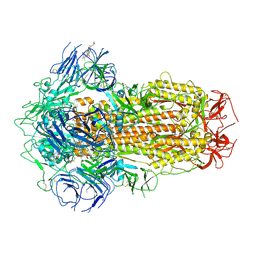 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
3UJL
 
 | | Crystal structure of abscisic acid bound PYL2 in complex with type 2C protein phosphatase ABI2 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
3UJG
 
 | | Crystal structure of SnRK2.6 in complex with HAB1 | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C 16, SULFATE ION, ... | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
3UJK
 
 | | Crystal structure of protein phosphatase ABI2 | | Descriptor: | MAGNESIUM ION, Protein phosphatase 2C 77 | | Authors: | Zhou, X.E, Soon, F.-F, Ng, L.-M, Kovach, A, Tan, M.H.E, Suino-Powell, K.M, He, Y, Xu, Y, Brunzelle, J.S, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2011-11-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases.
Science, 335, 2012
|
|
6V85
 
 | | Parainfluenza virus 5 L-P complex | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Abdella, R, He, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VAG
 
 | | Crystal structure of the oligomerization domain of phosphoprotein from parainfluenza virus 5 | | Descriptor: | GLYCEROL, Phosphoprotein | | Authors: | Aggarwal, M, Abdella, R, He, Y, Lamb, R.A. | | Deposit date: | 2019-12-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8HBC
 
 | | Crystal structure of the CysR-CTLD3 fragment of human DEC205 | | Descriptor: | Lymphocyte antigen 75 | | Authors: | Kong, D, Yu, B, Hu, Z, Cheng, C, Cao, L, He, Y. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Interaction of human dendritic cell receptor DEC205/CD205 with keratins.
J.Biol.Chem., 300, 2024
|
|
6V86
 
 | |
8ZNZ
 
 | | CD73 bound with HB0045 | | Descriptor: | 5'-nucleotidase, CALCIUM ION, HB0038 Fab heavy chain, ... | | Authors: | Xu, J, Wan, L, He, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | An antibody cocktail targeting two different CD73 epitopes enhances enzyme inhibition and tumor control.
Nat Commun, 15, 2024
|
|
1XU4
 
 | | ATPASE IN COMPLEX WITH AMP-PNP, MAGNESIUM AND POTASSIUM CO-F | | Descriptor: | DNA repair and recombination protein radA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Wu, Y, Qian, X, He, Y, Moya, I.A, Luo, Y. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence
J.Biol.Chem., 280, 2005
|
|
2LD7
 
 | | Solution structure of the mSin3A PAH3-SAP30 SID complex | | Descriptor: | Histone deacetylase complex subunit SAP30, Paired amphipathic helix protein Sin3a | | Authors: | Xie, T, He, Y, Korkeamaki, H, Zhang, Y, Imhoff, R, Lohi, O, Radhakrishnan, I. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the 30-kDa Sin3-associated protein (SAP30) in complex with the mammalian Sin3A corepressor and its role in nucleic acid binding.
J.Biol.Chem., 286, 2011
|
|
2KQ1
 
 | | Solution Structure Of Protein BH0266 From Bacillus halodurans. Northeast Structural Genomics Consortium Target BhR97a | | Descriptor: | BH0266 protein | | Authors: | Wu, Y, Eletsky, A, Lee, D, Sathyamoorthy, B, Buchwald, W, Hua, J, Ciccosanti, C, He, Y, Hamilton, K, Acton, T, Xiao, R, Everett, J, Lee, H, Prestegard, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein BH0266 From Bacillus halodurans
To be Published
|
|
