4LI8
 
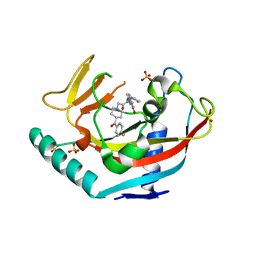 | | TANKYRASE-1 complexed with small molecule inhibitor 2-[4-(4-fluorobenzoyl)piperidin-1-yl]-N-[(4-oxo-3,5,7,8-tetrahydro-4H-pyrano[4,3-d]pyrimidin-2-yl)methyl]-N-(thiophen-2-ylmethyl)acetamide | | Descriptor: | 2-[4-(4-fluorobenzoyl)piperidin-1-yl]-N-[(4-oxo-3,5,7,8-tetrahydro-4H-pyrano[4,3-d]pyrimidin-2-yl)methyl]-N-(thiophen-2-ylmethyl)acetamide, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor.
J.Med.Chem., 56, 2013
|
|
7VAA
 
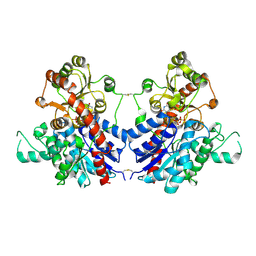 | |
7VA8
 
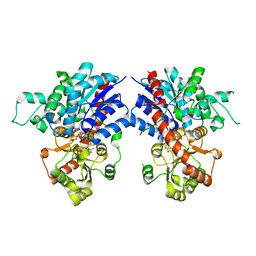 | | Crystal structure of MiCGT | | Descriptor: | UDP-glycosyltransferase 13, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhong, L, Zhang, Z.M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85003233 Å) | | Cite: | Directed Evolution of a Plant Glycosyltransferase for Chemo- and Regioselective Glycosylation of Pharmaceutically Significant Flavonoids
Acs Catalysis, 11, 2021
|
|
4G1A
 
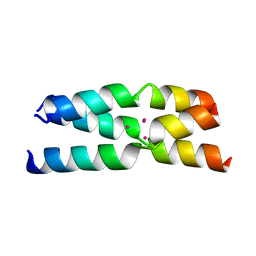 | |
4NC4
 
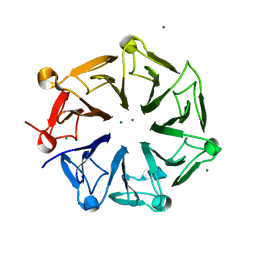 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4NBM
 
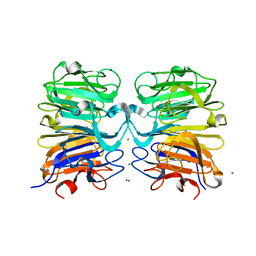 | | Crystal structure of UVB photoreceptor UVR8 and light-induced structural changes at 180K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
4Q2M
 
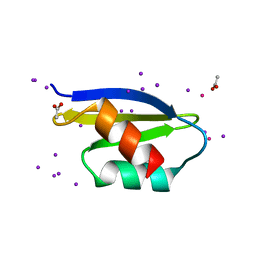 | |
4Q2L
 
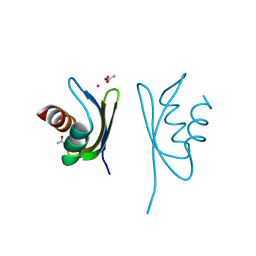 | |
4Q46
 
 | |
4NAA
 
 | | Crystal structure of UVB photoreceptor UVR8 from Arabidopsis thaliana and UV-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Ren, Z, Zhao, K.H. | | Deposit date: | 2013-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
7X5V
 
 | |
7DAT
 
 | | The crystal structure of COVID-19 main protease treated by AF | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAV
 
 | | The native crystal structure of COVID-19 main protease | | Descriptor: | COVID-19 MAIN PROTEASE | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAU
 
 | | The crystal structure of COVID-19 main protease treated by GA | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7YOC
 
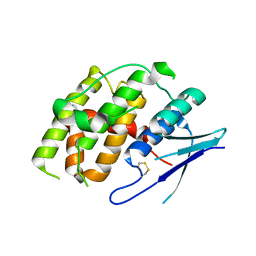 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2008-11-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
7WW2
 
 | | Structure of an Isocytosine specific deaminase Vcz | | Descriptor: | 8-oxoguanine deaminase, ZINC ION | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
7X1R
 
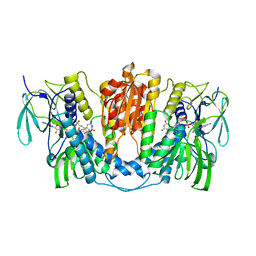 | | Cryo-EM structure of human thioredoxin reductase bound by Au | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GOLD ION, Thioredoxin reductase 1, ... | | Authors: | He, Z.S, Cao, P, Cao, S.H, He, B, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2022-02-24 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Au4 cluster inhibits human thioredoxin reductase activity via specifically binding of Au to Cys189
Nano Today, 47, 2022
|
|
6W89
 
 | | Structure of DNMT3A (R882H) in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), CITRIC ACID, DNA (cytosine-5)-methyltransferase 3-like, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W8J
 
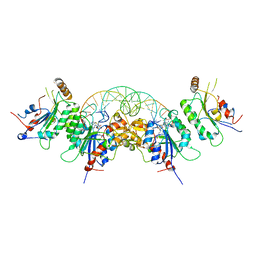 | | Structure of DNMT3A (R882H) in complex with CAG DNA | | Descriptor: | CAG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W8D
 
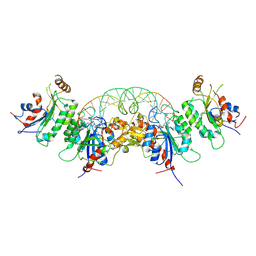 | | Structure of DNMT3A (R882H) in complex with CGT DNA | | Descriptor: | CGT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6YA5
 
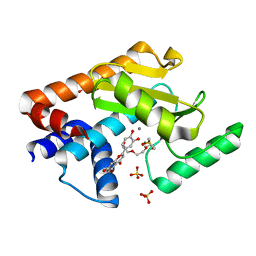 | | 2009 H1N1 PA Endonuclease in complex with LU2 | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unraveling the anti-influenza effect of flavonoids: Experimental validation of luteolin and its congeners as potent influenza endonuclease inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
8AB5
 
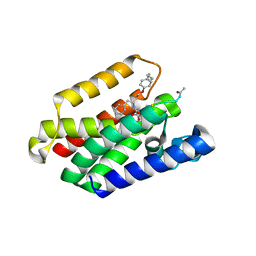 | | Structure of E. coli GlpG in complex with peptide derived inhibitor Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl] | | Descriptor: | Ac-VRHA-conh-[4-(4-butyl)-phenoxy-1-phenyl-2-butyl], Rhomboid protease GlpG | | Authors: | Skerlova, J, Polovinkin, V, Bach, K, Borshchevskiy, V, Strisovsky, K. | | Deposit date: | 2022-07-04 | | Release date: | 2023-10-25 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Extensive targeting of chemical space at the prime side of ketoamide inhibitors of rhomboid proteases by branched substituents empowers their selectivity and potency.
Eur.J.Med.Chem., 275, 2024
|
|
6WCZ
 
 | | CryoEM structure of full-length ZIKV NS5-hSTAT2 complex | | Descriptor: | Non-structural protein 5, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Boxiao, W, Stephanie, T, Kang, Z, Maria, T.S, Jian, F, Jiuwei, L, Linfeng, G, Wendan, R, Yanxiang, C, Ethan, C.V, HeaJin, H, Matthew, J.E, Sean, E.O, Adolfo, G.S, Hong, Z, Rong, H, Jikui, S. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for STAT2 suppression by flavivirus NS5.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YEM
 
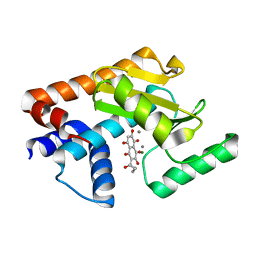 | | H1N1 2009 PA Endonuclease in complex with Quambalarine B | | Descriptor: | 3,5,6,8-tetrakis(oxidanyl)-2-pentanoyl-naphthalene-1,4-dione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Radilova, K, Brynda, J. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unraveling the anti-influenza effect of flavonoids: Experimental validation of luteolin and its congeners as potent influenza endonuclease inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
