7K7A
 
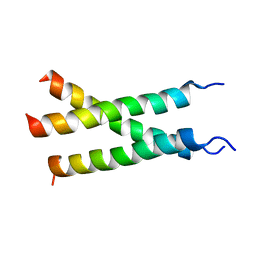 | | Transmembrane structure of TNFR1 | | Descriptor: | Tumor necrosis factor receptor superfamily member 1A | | Authors: | Zhao, L, Chou, J. | | Deposit date: | 2020-09-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Diversity and Similarity of Transmembrane Trimerization of TNF Receptors.
Front Cell Dev Biol, 8, 2020
|
|
3V6H
 
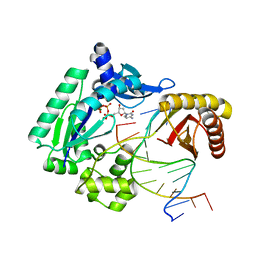 | | Replication of N2,3-Ethenoguanine by DNA Polymerases | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*G*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*(DOC))-3'), ... | | Authors: | Zhao, L. | | Deposit date: | 2011-12-19 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Replication of n(2) ,3-ethenoguanine by DNA polymerases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3V6K
 
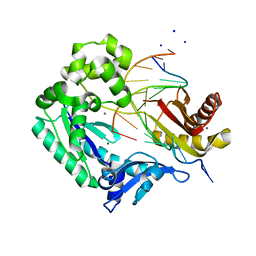 | | Replication of N2,3-Ethenoguanine by DNA Polymerases | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*AP*CP*(EFG)P*GP*AP*AP*TP*CP*CP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*GP*GP*AP*TP*TP*CP*(2DT))-3'), ... | | Authors: | Zhao, L. | | Deposit date: | 2011-12-20 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Replication of n(2) ,3-ethenoguanine by DNA polymerases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3V6J
 
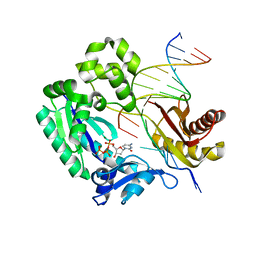 | | Replication of N2,3-Ethenoguanine by DNA Polymerases | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*(DOC))-3'), DNA (5'-D(*TP*CP*AP*TP*(EFG)P*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*C)-3'), ... | | Authors: | Zhao, L. | | Deposit date: | 2011-12-19 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Replication of n(2) ,3-ethenoguanine by DNA polymerases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4NOM
 
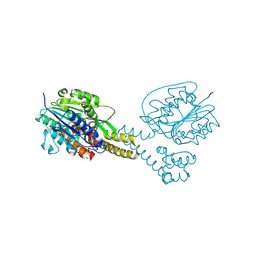 | | Crystal structure of asparaginyl endopeptidase (AEP)/Legumain activated at pH 4.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOK
 
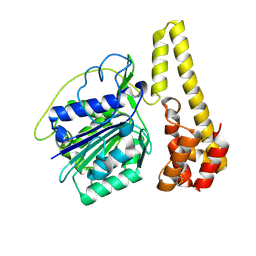 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOJ
 
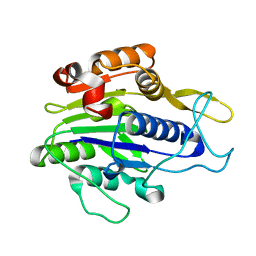 | | Crystal structure of the mature form of asparaginyl endopeptidase (AEP)/Legumain activated at pH 3.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOL
 
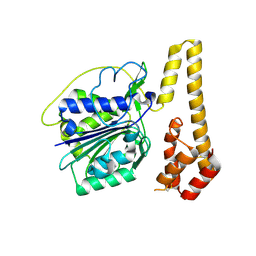 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain mutant D233A at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4FS1
 
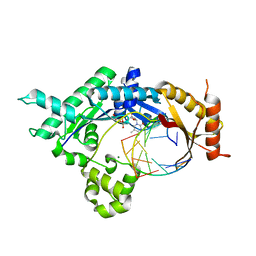 | | Base pairing mechanism of N2,3-ethenoguanine with dTTP by human polymerase iota | | Descriptor: | DNA 5'-D(*TP*CP*TP*(EFG)P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3', DNA polymerase iota, MAGNESIUM ION, ... | | Authors: | Zhao, L. | | Deposit date: | 2012-06-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Basis of Miscoding of the DNA Adduct N2,3-Ethenoguanine by Human Y-family DNA Polymerases.
J.Biol.Chem., 287, 2012
|
|
4FS2
 
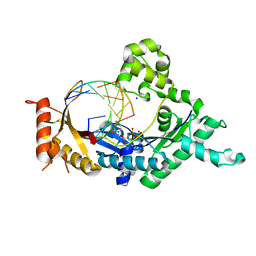 | | Base pairing mechanism of N2,3-ethenoguanine with dCTP by human polymerase iota | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*TP*CP*TP*(EFG)P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3'), DNA polymerase iota, ... | | Authors: | Zhao, L. | | Deposit date: | 2012-06-26 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Basis of Miscoding of the DNA Adduct N2,3-Ethenoguanine by Human Y-family DNA Polymerases.
J.Biol.Chem., 287, 2012
|
|
6ZP8
 
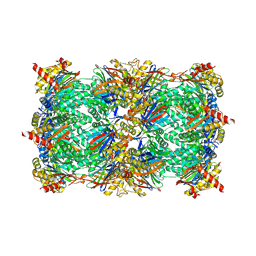 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB335 | | Descriptor: | (2~{S},3~{R})-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-2-(3-phenylpropanoylamino)butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
6ZOU
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB333 | | Descriptor: | 11-methyl-~{N}-[(2~{S},3~{R})-1-[[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]dodecanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
6ZP6
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB334 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
2OST
 
 | | The structure of a bacterial homing endonuclease : I-Ssp6803I | | Descriptor: | CALCIUM ION, Putative endonuclease, Synthetic DNA 29 MER | | Authors: | Zhao, L, Bonocora, R.P, Shub, D.A, Stoddard, B.L. | | Deposit date: | 2007-02-06 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif.
Embo J., 26, 2007
|
|
5ZC3
 
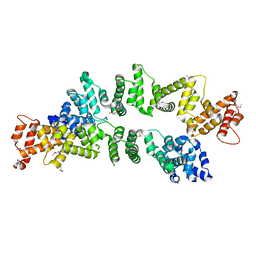 | | The Crystal Structure of PcRxLR12 | | Descriptor: | RxLR effector | | Authors: | Zhao, L, Zhang, X, Zhu, C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Crystal structure of the RxLR effector PcRxLR12 from Phytophthora capsici
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6L35
 
 | | PSI-LHCI Supercomplex from Physcometrella patens | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, L, Yan, Q.J, Qin, X.C. | | Deposit date: | 2019-10-09 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Antenna arrangement and energy-transfer pathways of PSI-LHCI from the moss Physcomitrella patens.
Cell Discov, 7, 2021
|
|
4TQS
 
 | |
4TQR
 
 | |
6K40
 
 | | Crystal structure of alkyl hydroperoxide reductase from D. radiodurans R1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkyl hydroperoxide reductase AhpD, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, M.-K, Zhang, J, Zhao, L. | | Deposit date: | 2019-05-22 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of the AhpD-like protein DR1765 from Deinococcus radiodurans R1.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
7WDK
 
 | | The structure of PldA-PA3488 complex | | Descriptor: | Phospholipase D, Tli4_C domain-containing protein | | Authors: | Zhao, L, Yang, X.Y, Li, Z.Q. | | Deposit date: | 2021-12-21 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into PA3488-mediated inactivation of Pseudomonas aeruginosa PldA
Nat Commun, 13, 2022
|
|
7W6L
 
 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6A
 
 | | Crystal structure of the MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L complex | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6J
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me2 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
