1LE2
 
 | |
5URO
 
 | | Structure of a soluble epoxide hydrolase identified in Trichoderma reesei | | Descriptor: | 2,2'-oxydi(ethyn-1-ol), Predicted protein | | Authors: | Wilson, C, de Oliveira, G, Chambergo, F, Dias, M.V.B. | | Deposit date: | 2017-02-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure of a soluble epoxide hydrolase identified in Trichoderma reesei.
Biochim. Biophys. Acta, 1865, 2017
|
|
1LPE
 
 | |
4CSV
 
 | | Tyrosine kinase AS - a common ancestor of Src and Abl bound to Gleevec | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, SRC-ABL TYROSINE KINASE ANCESTOR | | Authors: | Kutter, S, Wilson, C, Cruz, L, Agafonov, R.V, Hoemberger, M.S, Zorba, A, Kern, D. | | Deposit date: | 2014-03-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinase Dynamics. Using Ancient Protein Kinases to Unravel a Modern Cancer Drug'S Mechanism.
Science, 347, 2015
|
|
4UEU
 
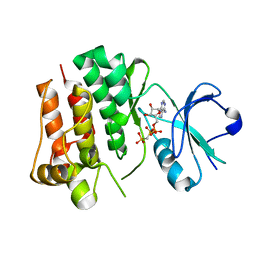 | | Tyrosine kinase AS - a common ancestor of Src and Abl | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, TYROSINE KINASE AS - A COMMON ANCESTOR OF SRC AND ABL | | Authors: | Kutter, S, Wilson, C, Agafonov, R.V, Hoemberger, M.S, Zorba, A, Halpin, J.C, Theobald, D.L, Kern, D. | | Deposit date: | 2014-12-18 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Kinase Dynamics. Using Ancient Protein Kinases to Unravel a Modern Cancer Drug'S Mechanism.
Science, 347, 2015
|
|
6UNW
 
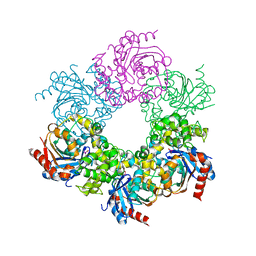 | | Epoxide hydrolase from an endophytic Streptomyces | | Descriptor: | CACODYLATE ION, CHLORIDE ION, Soluble epoxide hydrolase | | Authors: | Wilson, C, dos Santos, J.C, Dias, M.V.B. | | Deposit date: | 2019-10-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | An epoxide hydrolase from endophytic Streptomyces shows unique structural features and wide biocatalytic activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1LE4
 
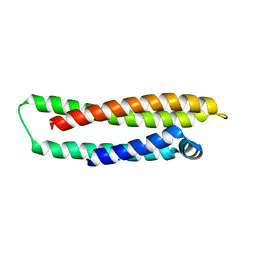 | |
1F1T
 
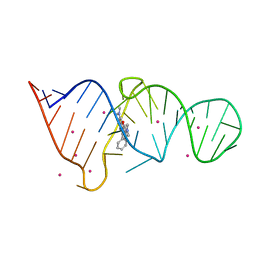 | | CRYSTAL STRUCTURE OF THE MALACHITE GREEN APTAMER COMPLEXED WITH TETRAMETHYL-ROSAMINE | | Descriptor: | MALACHITE GREEN APTAMER RNA, N,N'-TETRAMETHYL-ROSAMINE, STRONTIUM ION | | Authors: | Baugh, C, Grate, D, Wilson, C. | | Deposit date: | 2000-05-19 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A crystal structure of the malachite green aptamer.
J.Mol.Biol., 301, 2000
|
|
1DDY
 
 | |
1ET4
 
 | |
1F27
 
 | | CRYSTAL STRUCTURE OF A BIOTIN-BINDING RNA PSEUDOKNOT | | Descriptor: | BIOTIN, MAGNESIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*GP*UP*CP*CP*UP*C)-3'), ... | | Authors: | Nix, J, Sussman, D, Wilson, C. | | Deposit date: | 2000-05-23 | | Release date: | 2000-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a biotin-binding pseudoknot and the basis for RNA molecular recognition.
J.Mol.Biol., 296, 2000
|
|
7KBT
 
 | | Factor VIII in complex with the anti-C2 domain antibody, G99 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ronayne, E.K, Gish, J, Wilson, C, Peters, S, Spencer, H.T, Spiegel, P.C, Childers, K.C. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structure of Blood Coagulation Factor VIII in Complex With an Anti-C2 Domain Non-Classical, Pathogenic Antibody Inhibitor
Front Immunol, 12, 2021
|
|
4K4F
 
 | | Co-crystal structure of TNKS1 with compound 18 [4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide] | | Descriptor: | 4-[(4S)-5,5-dimethyl-2-oxo-4-phenyl-1,3-oxazolidin-3-yl]-N-(quinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X, Bregman, H, Wilson, C, DiMauro, E, Gunaydin, H. | | Deposit date: | 2013-04-12 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors.
J.Med.Chem., 56, 2013
|
|
1Q8N
 
 | | Solution Structure of the Malachite Green RNA Binding Aptamer | | Descriptor: | MALACHITE GREEN, RNA Aptamer | | Authors: | Flinders, J, DeFina, S.C, Brackett, D.M, Baugh, C, Wilson, C, Dieckmann, T. | | Deposit date: | 2003-08-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Recognition of planar and nonplanar ligands in the malachite green-RNA aptamer complex.
Chembiochem, 5, 2004
|
|
1J59
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | 5'-D(*AP*TP*AP*TP*GP*TP*CP*AP*CP*AP*CP*TP*TP*TP*TP*CP*G )-3', 5'-D(*GP*CP*GP*AP*AP*AP*AP*GP*TP*GP*TP*GP*AP*C)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Parkinson, G, Wilson, C, Gunasekera, A, Ebright, Y.W, Ebright, R.H, Berman, H.M. | | Deposit date: | 2002-03-01 | | Release date: | 2002-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface.
J.Mol.Biol., 260, 1996
|
|
7M7V
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with Compound 6 | | Descriptor: | 5-hydroxy-2-(4-hydroxyphenyl)-N-methyl-4-[(2-oxa-6-azaspiro[3.4]octan-6-yl)methyl]-1-benzofuran-3-carboxamide, Polyketide synthase Pks13 | | Authors: | Aggarwal, A, Sacchettini, J.C. | | Deposit date: | 2021-03-29 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Optimization of TAM16, a Benzofuran That Inhibits the Thioesterase Activity of Pks13; Evaluation toward a Preclinical Candidate for a Novel Antituberculosis Clinical Target.
J.Med.Chem., 65, 2022
|
|
5CEH
 
 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | Descriptor: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
8Q0U
 
 | | Identification and optimisation of novel inhibitors of the Polyketide synthetase 13 thioesterase domain with antitubercular activity | | Descriptor: | Polyketide synthase Pks13, SULFATE ION, ~{N}-[(1~{R})-2-[4-(azetidin-1-ylcarbonyl)phenyl]-1-cyano-ethyl]-3-(3,4-dimethoxyphenyl)-1,2,4-oxadiazole-5-carboxamide | | Authors: | Eadsforth, T.C, Punekar, A.S, Green, S.R, Baragana, B. | | Deposit date: | 2023-07-29 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Optimization of Novel Inhibitors of the Polyketide Synthase 13 Thioesterase Domain with Antitubercular Activity.
J.Med.Chem., 66, 2023
|
|
8Q17
 
 | | Identification and optimisation of novel inhibitors of the Polyketide synthetase 13 thioesterase domain with antitubercular activity | | Descriptor: | (2R)-2-{[(2R)-2-{[(2S)-2-{[(2R)-2-hydroxypropyl]oxy}propyl]oxy}propyl]oxy}propan-1-ol, GLYCEROL, Polyketide synthase Pks13, ... | | Authors: | Eadsforth, T.C, Punekar, A.S, Green, S.R, Baragana, B. | | Deposit date: | 2023-07-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Identification and Optimization of Novel Inhibitors of the Polyketide Synthase 13 Thioesterase Domain with Antitubercular Activity.
J.Med.Chem., 66, 2023
|
|
8Q0T
 
 | | Identification and optimisation of novel inhibitors of the Polyketide synthetase 13 thioesterase domain with antitubercular activity | | Descriptor: | 3-(3,4-dimethoxyphenyl)-~{N}-[2-(3,4-dimethoxyphenyl)ethyl]-1,2,4-oxadiazole-5-carboxamide, Polyketide synthase Pks13 | | Authors: | Eadsforth, T.C, Punekar, A.S, Green, S.R, Baragana, B. | | Deposit date: | 2023-07-29 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Optimization of Novel Inhibitors of the Polyketide Synthase 13 Thioesterase Domain with Antitubercular Activity.
J.Med.Chem., 66, 2023
|
|
5G40
 
 | | Crystal structure of adenylate kinase ancestor 4 with Zn and AMP-ADP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
8OS5
 
 | | Crystal structure of the Factor XII heavy chain reveals an interlocking dimer with a FnII to kringle domain interaction | | Descriptor: | Coagulation factor XII-Mie | | Authors: | Li, C, Saleem, M, Kaira, B.G, Brown, A, Wilson, C, Philippou, H, Emsley, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Factor XII and kininogen asymmetric assembly with gC1qR/C1QBP/P32 is governed by allostery.
Blood, 136, 2020
|
|
5G3Z
 
 | | Crystal structure of adenylate kinase ancestor 3 with Zn, Mg and Ap5A bound | | Descriptor: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G3Y
 
 | | Crystal structure of adenylate kinase ancestor 1 with Zn and ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINSE, ZINC ION | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
5G41
 
 | | Crystal structure of adenylate kinase ancestor 4 with Zn, Mg and Ap5A bound | | Descriptor: | ADENYLATE KINSE, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nguyen, V, Kutter, S, English, J, Kern, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evolutionary drivers of thermoadaptation in enzyme catalysis.
Science, 355, 2017
|
|
