4V5I
 
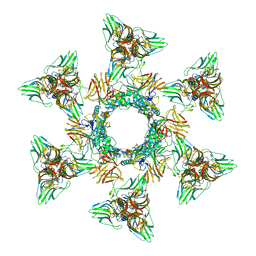 | | Structure of the Phage P2 Baseplate in its Activated Conformation with Ca | | Descriptor: | CALCIUM ION, ORF15, ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.464 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WZP
 
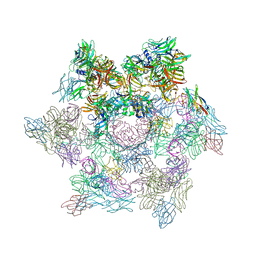 | | Structures of Lactococcal Phage p2 Baseplate Shed Light on a Novel Mechanism of Host Attachment and Activation in Siphoviridae | | Descriptor: | CAMELID VHH5, LACTOCOCCAL PHAGE P2 ORF15, LACTOCOCCAL PHAGE P2 ORF16, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2009-12-01 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1LQ9
 
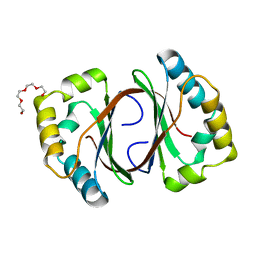 | | Crystal Structure of a Monooxygenase from the Gene ActVA-Orf6 of Streptomyces coelicolor Strain A3(2) | | Descriptor: | ACTVA-ORF6 MONOOXYGENASE, TETRAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
EMBO J., 22, 2003
|
|
2X53
 
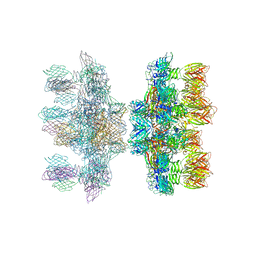 | | Structure of the phage p2 baseplate in its activated conformation with Sr | | Descriptor: | ORF15, ORF16, PUTATIVE RECEPTOR BINDING PROTEIN, ... | | Authors: | Sciara, G, Bebeacua, C, Bron, P, Tremblay, D, Ortiz-Lombardia, M, Lichiere, J, van Heel, M, Campanacci, V, Moineau, S, Cambillau, C. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1N5Q
 
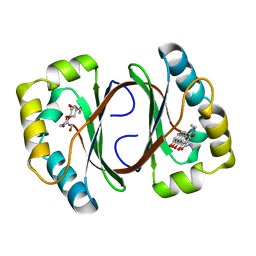 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with dehydrated Sancycline | | Descriptor: | 4-DIMETHYLAMINO-1,10,11,12-TETRAHYDROXY-3-OXO-3,4,4A,5-TETRAHYDRO-NAPHTHACENE-2-CARBOXYLIC ACID AMIDE, ActaVA-Orf6 monooxygenase, HEXAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5S
 
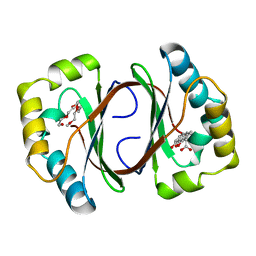 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9-OXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5T
 
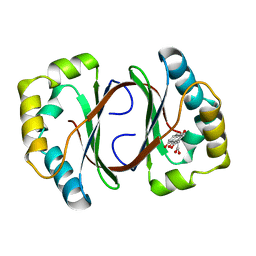 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Oxidized Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9,10-DIOXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, G Kendrew, S, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5V
 
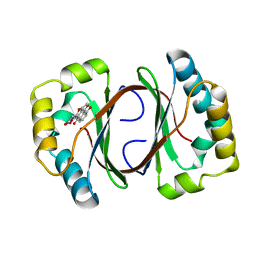 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Nanaomycin D | | Descriptor: | 7-HYDROXY-5-METHYL-3,3A,5,11B-TETRAHYDRO-1,4-DIOXA-CYCLOPENTA[A]ANTHRACENE-2,6,11-TRIONE, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
4O6M
 
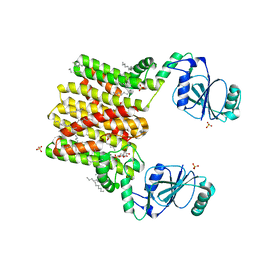 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CMP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4O6N
 
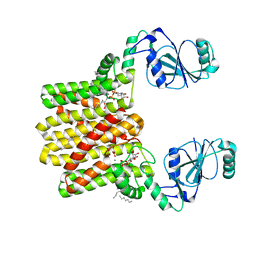 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CDP-bound) | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-12-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4Q7C
 
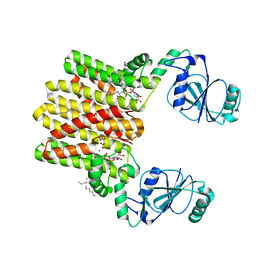 | | Structure of AF2299, a CDP-alcohol phosphotransferase | | Descriptor: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | Authors: | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
3GKO
 
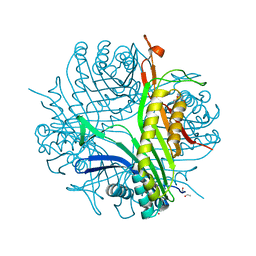 | | Crystal structure of urate oxydase using surfactant Poloxamer 188 as a New Crystallizing Agent | | Descriptor: | 8-AZAXANTHINE, POTASSIUM ION, Uricase | | Authors: | Delfosse, V, Giffard, M, Sciara, G, Bonnete, F, Mayer, C. | | Deposit date: | 2009-03-11 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Surfactant Poloxamer 188 as a New Crystallizing Agent for Urate Oxidase
Cryst.Growth Des., 9, 2009
|
|
6XUV
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, laminaribiose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6XUU
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, glucose-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Vallone, B, Sciara, G. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
6XUT
 
 | | Crystallographic structure of oligosaccharide dehydrogenase from Pycnoporus cinnabarinus, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cerutti, G, Savino, C, Montemiglio, L.C, Sciara, G, Vallone, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and functional characterization of an oligosaccharide dehydrogenase from Pycnoporus cinnabarinus provides insights into fungal breakdown of lignocellulose.
Biotechnol Biofuels, 14, 2021
|
|
3D9B
 
 | | Symmetric structure of E. coli AcrB | | Descriptor: | Acriflavine resistance protein B, NICKEL (II) ION | | Authors: | Veesler, D, Blangy, S, Cambillau, C, Sciara, G. | | Deposit date: | 2008-05-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | There is a baby in the bath water: AcrB contamination is a major problem in membrane-protein crystallization.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1K3G
 
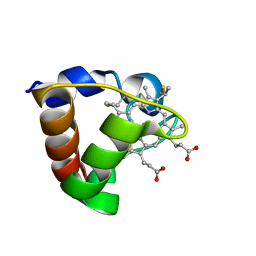 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3H
 
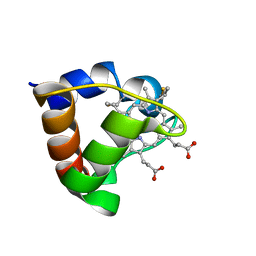 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
2JJN
 
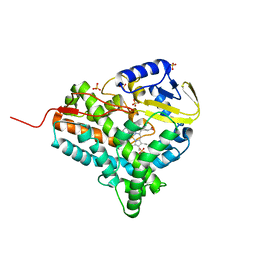 | | Structure of closed cytochrome P450 EryK | | Descriptor: | CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2JJO
 
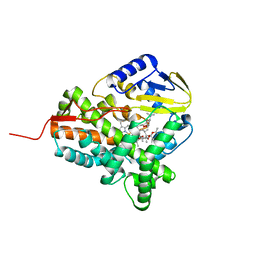 | | Structure of cytochrome P450 EryK in complex with its natural substrate erD | | Descriptor: | CYTOCHROME P450 113A1, Erythromycin D, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
7Z3B
 
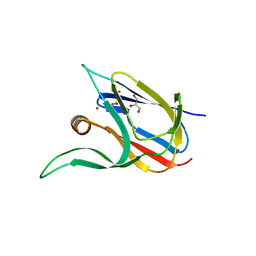 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, reduced form | | Descriptor: | ACETATE ION, AcoP, COPPER (I) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3F
 
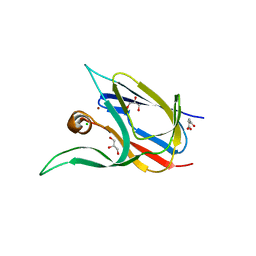 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, oxidized form | | Descriptor: | ACETATE ION, AcoP, CHLORIDE ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3G
 
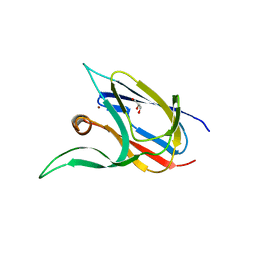 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, H166A mutant | | Descriptor: | AcoP, COPPER (I) ION, GLYCEROL | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3I
 
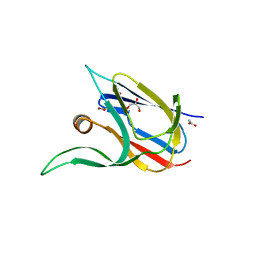 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, M171A mutant | | Descriptor: | ACETATE ION, AcoP, COPPER (II) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
2W72
 
 | | DEOXYGENATED STRUCTURE OF A DISTAL SITE HEMOGLOBIN MUTANT PLUS XE | | Descriptor: | HUMAN HEMOGLOBIN A, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Miele, A.E, Draghi, F, Sciara, G, Johnson, K.A, Renzi, F, Vallone, B, Brunori, M, Savino, C. | | Deposit date: | 2008-12-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Pattern of Cavities in Globins: The Case of Human Hemoglobin.
Biopolymers, 91, 2009
|
|
