1MWQ
 
 | | Structure of HI0828, a Hypothetical Protein from Haemophilus influenzae with a Putative Active-Site Phosphohistidine | | Descriptor: | CACODYLATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-30 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site
Proteins, 59, 2005
|
|
1JO0
 
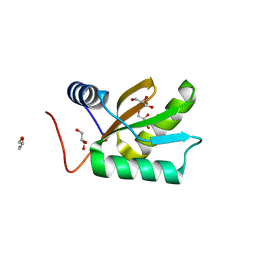 | | Structure of HI1333, a Hypothetical Protein from Haemophilus influenzae with Structural Similarity to RNA-binding Proteins | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN HI1333 | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae
Proteins, 49, 2002
|
|
2HPR
 
 | |
5IN4
 
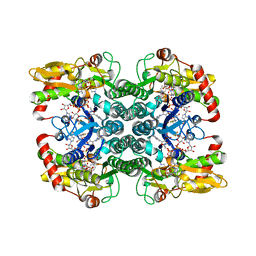 | | Crystal Structure of GDP-mannose 4,6 dehydratase bound to a GDP-fucose based inhibitor | | Descriptor: | GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sickmier, E.A. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Facile Modulation of Antibody Fucosylation with Small Molecule Fucostatin Inhibitors and Cocrystal Structure with GDP-Mannose 4,6-Dehydratase.
Acs Chem.Biol., 11, 2016
|
|
5IN5
 
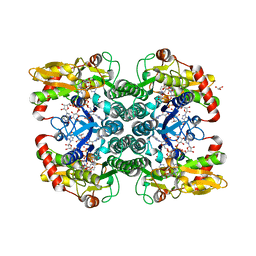 | |
1BKC
 
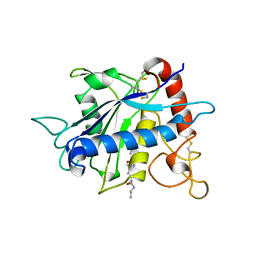 | | CATALYTIC DOMAIN OF TNF-ALPHA CONVERTING ENZYME (TACE) | | Descriptor: | N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-methyl-L-valyl-N-(2-aminoethyl)-L-alaninamide, TUMOR NECROSIS FACTOR-ALPHA-CONVERTING ENZYME, ZINC ION | | Authors: | Maskos, K, Fernandez-Catalan, C, Bode, W. | | Deposit date: | 1998-04-23 | | Release date: | 1999-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic domain of human tumor necrosis factor-alpha-converting enzyme.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1SPH
 
 | |
1GPR
 
 | |
1IN0
 
 | |
1DBF
 
 | |
1PCH
 
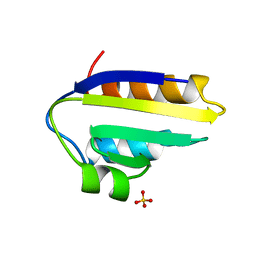 | |
6VGL
 
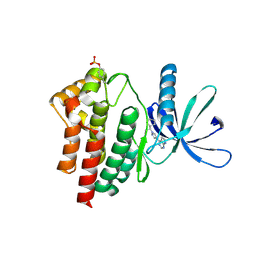 | | JAK2 JH1 in complex with ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNB
 
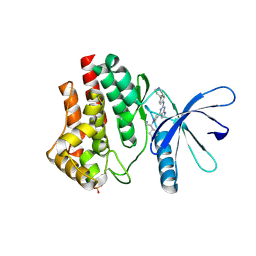 | | JAK2 JH1 in complex with BL2-084 | | Descriptor: | (3S)-3-cyclopentyl-3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNE
 
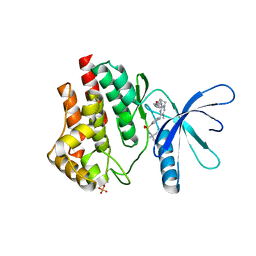 | | JAK2 JH1 in complex with Fedratinib | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNC
 
 | | JAK2 JH1 in complex with BL2-096 | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNG
 
 | | JAK2 JH1 in complex with PN2-118 | | Descriptor: | N-{2-fluoro-5-[(2-{[3-fluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]phenyl}-2-methylpropane-2-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNH
 
 | | JAK2 JH1 in complex with PN2-123 | | Descriptor: | N-{5-[(2-{[3,5-difluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]-2-fluorophenyl}-2-methylpropane-2-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNF
 
 | | JAK2 JH1 in complex with MA9-086 | | Descriptor: | N~4~-[1-(tert-butylsulfonyl)-2,3-dihydro-1H-indol-6-yl]-N~2~-[3-fluoro-4-(1-methylpiperidin-4-yl)phenyl]-5-methylpyrimidine-2,4-diamine, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VN8
 
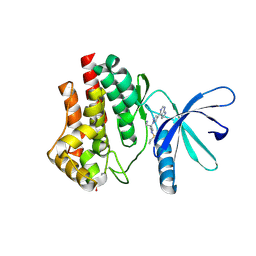 | | JAK2 JH1 in complex with baricitinib | | Descriptor: | Baricitinib, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNJ
 
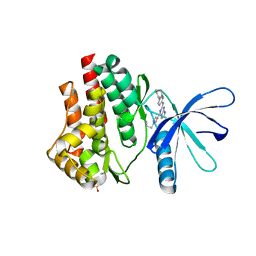 | | JAK2 JH1 in complex with PN4-014 | | Descriptor: | 3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNM
 
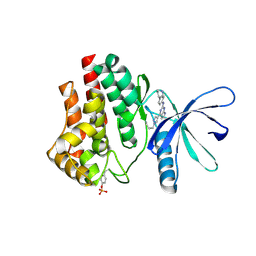 | | JAK2 JH1 in complex with SY5-103 | | Descriptor: | 4-[1-(but-3-en-1-yl)-1H-pyrazol-4-yl]-N-[4-(piperidin-4-yl)phenyl]-7H-pyrrolo[2,3-d]pyrimidin-2-amine, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNI
 
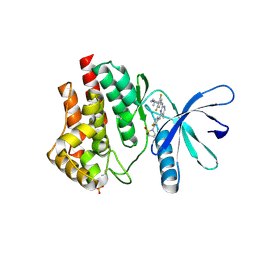 | | JAK2 JH1 in complex with PN3-115 | | Descriptor: | 2-{5-[(2-{[3,5-difluoro-4-(1-methylpiperidin-4-yl)phenyl]amino}-5-methylpyrimidin-4-yl)amino]-2-fluorophenyl}-1lambda~6~,2-thiazolidine-1,1-dione, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNL
 
 | | JAK2 JH1 in complex with SG3-179 | | Descriptor: | 4-[[4-[[3-(~{tert}-butylsulfonylamino)-4-chloranyl-phenyl]amino]-5-methyl-pyrimidin-2-yl]amino]-2-fluoranyl-~{N}-(1-methylpiperidin-4-yl)benzamide, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VNK
 
 | | JAK2 JH1 in complex with PN4-073 | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
6VS3
 
 | | JAK2 JH1 in complex with BL2-057 | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(2-{[4-(piperidin-4-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Tyrosine-protein kinase JAK2 | | Authors: | Davis, R.R, Schonbrunn, E. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into JAK2 Inhibition by Ruxolitinib, Fedratinib, and Derivatives Thereof.
J.Med.Chem., 64, 2021
|
|
