1MB6
 
 | |
8WCM
 
 | |
2I1T
 
 | |
9AZX
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with NDPr | | Descriptor: | Non-structural protein 3, {(2R,3S,4R,5R)-5-[(8S)-4-aminopyrrolo[2,1-f][1,2,4]triazin-7-yl]-5-cyano-3,4-dihydroxyoxolan-2-yl}methyl [(2R,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2024-03-11 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | GS-441524-Diphosphate-Ribose Derivatives as Nanomolar Binders and Fluorescence Polarization Tracers for SARS-CoV-2 and Other Viral Macrodomains.
Acs Chem.Biol., 19, 2024
|
|
2JOT
 
 | |
8RTW
 
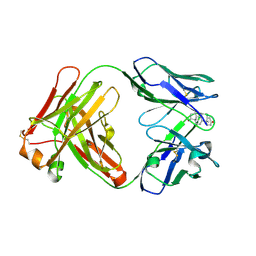 | | Crystal Structure of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | Anti-testosterone Fab 220 heavy chain, Anti-testosterone Fab 220 light chain, TESTOSTERONE | | Authors: | Eronen, V, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into ternary immunocomplex formation and cross-reactivity: binding of an anti-immunocomplex FabB12 to Fab220-testosterone complex.
Febs J., 291, 2024
|
|
8RTX
 
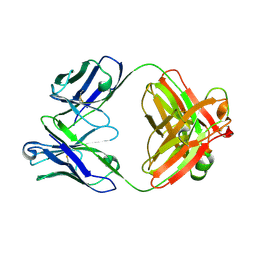 | | Crystal Structure of an Anti-idiotype Fab Fragment | | Descriptor: | Anti-IC Fab fragment B12 heavy chain, Anti-IC Fab fragment B12 light chain | | Authors: | Eronen, V, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into ternary immunocomplex formation and cross-reactivity: binding of an anti-immunocomplex FabB12 to Fab220-testosterone complex.
Febs J., 291, 2024
|
|
1Z2E
 
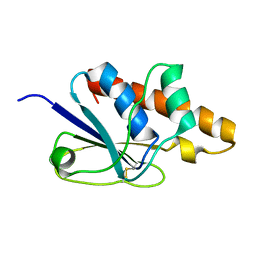 | | Solution Structure of Bacillus subtilis ArsC in oxidized state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
1Z2D
 
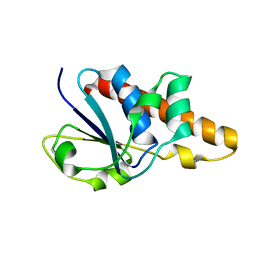 | | Solution Structure of Bacillus subtilis ArsC in reduced state | | Descriptor: | Arsenate reductase | | Authors: | Jin, C, Li, Y. | | Deposit date: | 2005-03-08 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures and Backbone Dynamics of Arsenate Reductase from Bacillus subtilis: REVERSIBLE CONFORMATIONAL SWITCH ASSOCIATED WITH ARSENATE REDUCTION
J.Biol.Chem., 280, 2005
|
|
3R1G
 
 | | Structure Basis of Allosteric Inhibition of BACE1 by an Exosite-Binding Antibody | | Descriptor: | Beta-secretase 1, FAB of YW412.8.31 antibody heavy chain, FAB of YW412.8.31 antibody light chain | | Authors: | Wang, W, Rouge, L, Wu, P, Chiu, C, Chen, Y, Wu, Y, Watts, R.J. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Therapeutic Antibody Targeting BACE1 Inhibits Amyloid-{beta} Production in Vivo.
Sci Transl Med, 3, 2011
|
|
8GIA
 
 | | Crystal structure of SARS-CoV-2 (Covid-19) Nsp3 macrodomain in complex with TFMU-ADPr | | Descriptor: | Non-structural protein 3, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl [(2R,3S,4R,5R)-3,4-dihydroxy-5-{[2-oxo-4-(trifluoromethyl)-2H-1-benzopyran-7-yl]oxy}oxolan-2-yl]methyl dihydrogen diphosphate | | Authors: | Wallace, S.D, Bagde, S.R, Fromme, J.C. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Fluorescence Polarization Assay for Macrodomains Facilitates the Identification of Potent Inhibitors of the SARS-CoV-2 Macrodomain.
Acs Chem.Biol., 18, 2023
|
|
7YSX
 
 | | Crystal structure of PDE4D complexed with licoisoflavone A | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-2,4-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YQF
 
 | | Crystal structure of PDE4D complexed with glycyrrhisoflavone | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-(3-methylbut-2-enyl)-4,5-bis(oxidanyl)phenyl]-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2022-08-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Bioactive compounds from Huashi Baidu decoction possess both antiviral and anti-inflammatory effects against COVID-19.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7MYJ
 
 | | Structure of full length human AMPK (a2b1g1) in complex with a small molecule activator MSG011 | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Ovens, A.J, Gee, Y.S, Ling, N.X.Y, Waters, N.J, Yu, D, Scott, J.W, Parker, M.W, Hoffman, N.J, Kemp, B.E, Baell, J.B, Oakhill, J.S, Langendorf, C.G. | | Deposit date: | 2021-05-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-function analysis of the AMPK activator SC4 and identification of a potent pan AMPK activator.
Biochem.J., 479, 2022
|
|
