4WF1
 
 | | Crystal structure of the E. coli ribosome bound to negamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Olivier, N.B, Altman, R.B, Noeske, J, Basarab, G.S, Code, E, Ferguson, A.D, Gao, N, Huang, J, Juette, M.F, Livchak, S, Miller, M.D, Prince, D.B, Cate, J.H.D, Buurman, E.T, Blanchard, S.C. | | Deposit date: | 2014-09-11 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Negamycin induces translational stalling and miscoding by binding to the small subunit head domain of the Escherichia coli ribosome.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3BSW
 
 | |
3BSY
 
 | |
3BSS
 
 | | PglD from Campylobacter jejuni, NCTC 11168, with native substrate | | Descriptor: | Acetyltransferase, UDP-2-acetamido-4-amino-2,4,6-trideoxy-alpha-D-glucopyranose | | Authors: | Olivier, N.B, Imperiali, B. | | Deposit date: | 2007-12-26 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and catalytic mechanism of PglD from Campylobacter jejuni.
J.Biol.Chem., 283, 2008
|
|
5VFD
 
 | | Diazabicyclooctenone ETX2514 bound to Class D beta lactamase OXA-24 from A. baumannii | | Descriptor: | (2S,5R)-1-formyl-4-methyl-5-[(sulfooxy)amino]-1,2,5,6-tetrahydropyridine-2-carboxamide, (2S,5R)-4-methyl-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]oct-3-ene-2-carboxamide, Beta-lactamase, ... | | Authors: | Olivier, N.B, Lahiri, S. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | ETX2514 is a broad-spectrum beta-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii.
Nat Microbiol, 2, 2017
|
|
4XWA
 
 | | TMK from S.aureus in complex with the Piperidinyl Thymine class inhibitor with a C5 ethyl-amine | | Descriptor: | 2-(3-chlorophenoxy)-6-(ethylamino)-4-[(R)-[(3S)-3-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)piperidin-1-yl](phenyl)methyl]benzoic acid, Thymidylate kinase | | Authors: | Olivier, N.B. | | Deposit date: | 2015-01-28 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A highly potent antibacterial inhibitor of Gram-positive bacterial thymidylate kinase (TMK): SAR of piperidinyl thymines at position C5 and L1
Bioorg Med Chem Lett, 2015
|
|
3FFQ
 
 | | HCN2I 443-640 apo-state | | Descriptor: | BROMIDE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Olivier, N.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the structure and conformational movements of proteins with transition metal ion FRET.
Nat.Methods, 6, 2009
|
|
4HDC
 
 | |
4GFD
 
 | | Thymidylate kinase (TMK) from S. Aureus in complex with TK-666 | | Descriptor: | 2-(3-bromophenoxy)-4-{(1R)-3,3-dimethyl-1-[(3S)-3-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)piperidin-1-yl]butyl}benzoic acid, Thymidylate kinase | | Authors: | Olivier, N.B, Martinez-Botella, G, Keating, T. | | Deposit date: | 2012-08-03 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In Vivo Validation of Thymidylate Kinase (TMK) with a Rationally Designed, Selective Antibacterial Compound.
Acs Chem.Biol., 7, 2012
|
|
4OKG
 
 | |
4OZE
 
 | | A.aolicus LpxC in complex with native product | | Descriptor: | CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION, ... | | Authors: | Olivier, N.B. | | Deposit date: | 2014-02-15 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mechanistic insight from the crystal structure of A.aolicus LpxC in the presence of product
To Be Published
|
|
4QG7
 
 | |
4QGA
 
 | |
4QGH
 
 | | S.aureus TMK in complex with potent inhibitor compound 47 | | Descriptor: | 2-(3-chlorophenoxy)-3-fluoro-4-{(1S)-3-methyl-1-[(3S)-3-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)piperidin-1-yl]butyl}benzoic acid, Thymidylate kinase | | Authors: | Olivier, N.B. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Antibacterial inhibitors of gram-positive thymidylate kinase: structure-activity relationships and chiral preference of a new hydrophobic binding region.
J.Med.Chem., 57, 2014
|
|
4QGF
 
 | |
4QGG
 
 | |
3JXT
 
 | |
3NU8
 
 | |
3NUB
 
 | | WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with Product as the External Aldimine | | Descriptor: | (2S,3S,4R,5R,6R)-5-(acetylamino)-6-{[(S)-{[(S)-{[(2S,3R,4S,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-3-hydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}tetrahydro-2H-pyran-2-carboxylic acid, Aminotransferase WbpE, GLYCEROL, ... | | Authors: | Larkin, A, Olivier, N.B, Imperiali, B. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of WbpE from Pseudomonas aeruginosa PAO1: A Nucleotide Sugar Aminotransferase Involved in O-Antigen Assembly
Biochemistry, 49, 2010
|
|
3NU7
 
 | | WbpE, an Aminotransferase from Pseudomonas aeruginosa Involved in O-antigen Assembly in Complex with the Cofactor PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase WbpE, GLYCEROL | | Authors: | Larkin, A, Olivier, N.B, Imperiali, B. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Analysis of WbpE from Pseudomonas aeruginosa PAO1: A Nucleotide Sugar Aminotransferase Involved in O-Antigen Assembly
Biochemistry, 49, 2010
|
|
3GSL
 
 | |
4GSY
 
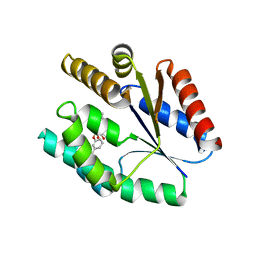 | |
1Q43
 
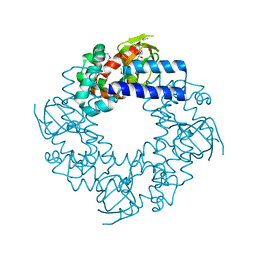 | | HCN2I 443-640 in the presence of cAMP, selenomethionine derivative | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-08-01 | | Release date: | 2003-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for modulation and agonist specificity of HCN pacemaker channels
Nature, 425, 2003
|
|
1Q3E
 
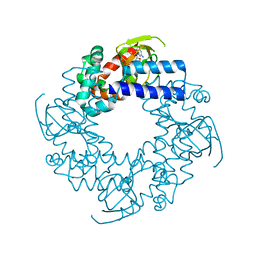 | | HCN2J 443-645 in the presence of cGMP | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for modulation and agonist specificity of HCN pacemaker channels
Nature, 425, 2003
|
|
1Q5O
 
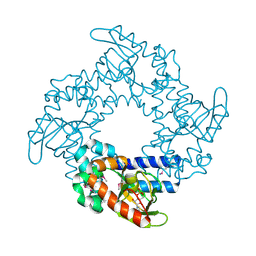 | | HCN2J 443-645 in the presence of cAMP, selenomethionine derivative | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-08-08 | | Release date: | 2003-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | STRUCTURAL BASIS FOR MODULATION AND AGONIST SPECIFICITY OF HCN PACEMAKER CHANNELS
Nature, 425, 2003
|
|
