4TNM
 
 | |
5NGO
 
 | |
7PLQ
 
 | | Crystal structure of the PARP domain of wheat SRO1 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Wirthmueller, L, Loll, B. | | Deposit date: | 2021-09-01 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The superior salinity tolerance of bread wheat cultivar Shanrong No. 3 is unlikely to be caused by elevated Ta-sro1 poly-(ADP-ribose) polymerase activity.
Plant Cell, 34, 2022
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1PFL
 
 | | REFINED SOLUTION STRUCTURE OF HUMAN PROFILIN I | | Descriptor: | PROFILIN I | | Authors: | Metzler, W.J, Farmer II, B.T, Constantine, K.L, Friedrichs, M.S, Lavoie, T, Mueller, L. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of human profilin I.
Protein Sci., 4, 1995
|
|
4TGF
 
 | | SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA | | Descriptor: | DES-VAL-1,VAL-2,TRANSFORMING GROWTH FACTOR, ALPHA | | Authors: | Kline, T.P, Brown, F.K, Brown, S.C, Jeffs, P.W, Kopple, K.D, Mueller, L. | | Deposit date: | 1990-06-13 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human transforming growth factor alpha derived from 1H NMR data.
Biochemistry, 29, 1990
|
|
3GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
4GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
3KJ6
 
 | | Crystal structure of a Methylated beta2 Adrenergic Receptor-Fab complex | | Descriptor: | Beta-2 adrenergic receptor, Fab heavy chain, Fab light chain, ... | | Authors: | Bokoch, M.P, Zou, Y, Rasmussen, S.G.F, Liu, C.W, Nygaard, R, Rosenbaum, D.M, Fung, J.J, Choi, H.-J, Thian, F.S, Kobilka, T.S, Puglisi, J.D, Weis, W.I, Pardo, L, Prosser, S, Mueller, L, Kobilka, B.K. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor.
Nature, 463, 2010
|
|
1AH1
 
 | | CTLA-4, NMR, 20 STRUCTURES | | Descriptor: | CTLA-4, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Metzler, W.J, Bajorath, J, Fenderson, W, Shaw, S.-Y, Peach, R, Constantine, K.L, Naemura, J, Leytze, G, Lavoie, T.B, Mueller, L, Linsley, P.S. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human CTLA-4 and delineation of a CD80/CD86 binding site conserved in CD28.
Nat.Struct.Biol., 4, 1997
|
|
1AKP
 
 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
1BLJ
 
 | | NMR ENSEMBLE OF BLK SH2 DOMAIN, 20 STRUCTURES | | Descriptor: | P55 BLK PROTEIN TYROSINE KINASE | | Authors: | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | Deposit date: | 1996-03-26 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
2GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1GBR
 
 | | ORIENTATION OF PEPTIDE FRAGMENTS FROM SOS PROTEINS BOUND TO THE N-TERMINAL SH3 DOMAIN OF GRB2 DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, SOS-A PEPTIDE | | Authors: | Wittekind, M, Mapelli, C, Farmer, B.T, Suen, K.-L, Goldfarb, V, Tsao, J, Lavoie, T, Barbacid, M, Meyers, C.A, Mueller, L. | | Deposit date: | 1994-08-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1BLK
 
 | | NMR ENSEMBLE OF BLK SH2 DOMAIN USING CHEMICAL SHIFT REFINEMENT, 20 STRUCTURES | | Descriptor: | P55 BLK PROTEIN TYROSINE KINASE | | Authors: | Metzler, W.J, Leiting, B, Pryor, K, Mueller, L, Farmer II, B.T. | | Deposit date: | 1996-03-26 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the SH2 domain from p55blk kinase.
Biochemistry, 35, 1996
|
|
1NF1
 
 | | THE GAP RELATED DOMAIN OF NEUROFIBROMIN | | Descriptor: | PROTEIN (NEUROFIBROMIN) | | Authors: | Scheffzek, K, Ahmadian, M.R, Wiesmueller, L, Kabsch, W, Stege, P, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1998-07-08 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the GAP-related domain from neurofibromin and its implications.
EMBO J., 17, 1998
|
|
6OSO
 
 | | The crystal structure of the isolate tryptophan synthase alpha-chain from Salmonella enterica serovar typhimurium at 1.75 Angstrom resolution | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L, Chang, C, Fan, L. | | Deposit date: | 2019-05-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Backbone assignments and conformational dynamics in the S. typhimurium tryptophan synthase alpha-subunit from solution-state NMR.
J.Biomol.Nmr, 74, 2020
|
|
6OUY
 
 | | The crystal structure of the isolate tryptophan synthase alpha-chain from Salmonella enterica serovar typhimurium at 1.60 Angstrom resolution | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L, Chang, C, Fan, L. | | Deposit date: | 2019-05-06 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Backbone assignments and conformational dynamics in the S. typhimurium tryptophan synthase alpha-subunit from solution-state NMR.
J.Biomol.Nmr, 74, 2020
|
|
5K4J
 
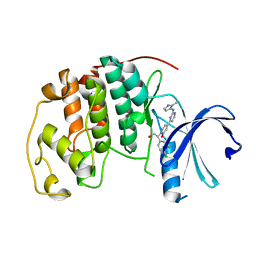 | | Crystal Structure of CDK2 in complex with compound 22 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Cyclin-dependent kinase 2 | | Authors: | Yin, J, Wang, W. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an Extracellular Signal-Regulated Kinase 1/2 (ERK1/2) Inhibitor in Early Clinical Development.
J.Med.Chem., 59, 2016
|
|
9CUB
 
 | | Human STING G230A/R293Q variant bound to diABZI-a1 | | Descriptor: | 1,1'-[(2E)-but-2-ene-1,4-diyl]bis{2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-1,3-benzimidazole-5-carboxamide}, Stimulator of interferon genes protein | | Authors: | Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING
To Be Published
|
|
9CUA
 
 | |
9CUD
 
 | | Human STING G230A/R293Q variant bound to diABZI-i | | Descriptor: | (2E)-1-[(2E)-4-{(2E)-5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)imino]-3-methyl-2,3-dihydro-1H-1,3-benzimidazol-1-yl}-2,3-dimethylbut-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)imino]-7-[(3-methoxyphenyl)methoxy]-3-methyl-2,3-dihydro-1H-1,3-benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING
To Be Published
|
|
9CUE
 
 | | Human STING H232R variant bound to ABZI | | Descriptor: | Stimulator of interferon genes protein, benzyl (3-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methoxy-1H-1,3-benzimidazol-1-yl}propyl)carbamate | | Authors: | Sack, J.S, Critton, D.A. | | Deposit date: | 2024-07-26 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Orthosteric STING inhibition elucidates molecular correction of SAVI STING
To Be Published
|
|
