3W95
 
 | |
5YBH
 
 | |
5YK3
 
 | | human Ragulator complex | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Wu, G, Mu, Z. | | Deposit date: | 2017-10-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane
Cell Discov, 3, 2017
|
|
5YK5
 
 | | structure of the human Lamtor4-Lamtor5 complex | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5 | | Authors: | Wu, G, Mu, Z. | | Deposit date: | 2017-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane
Cell Discov, 3, 2017
|
|
5JMT
 
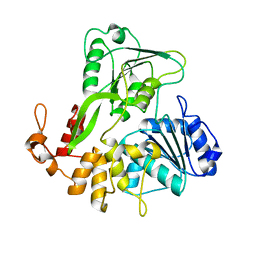 | | Crystal structure of Zika virus NS3 helicase | | Descriptor: | NS3 helicase | | Authors: | Tian, H, Ji, X, Yang, X, Xie, W, Yang, K, Chen, C, Wu, C, Chi, H, Mu, Z, Wang, Z, Yang, H. | | Deposit date: | 2016-04-29 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | The crystal structure of Zika virus helicase: basis for antiviral drug design
Protein Cell, 7, 2016
|
|
8IT0
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-2) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISZ
 
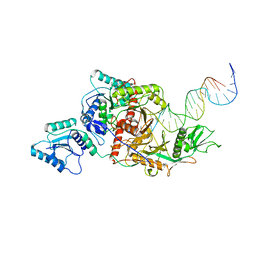 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
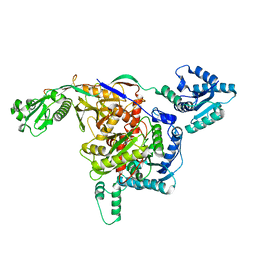 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IT1
 
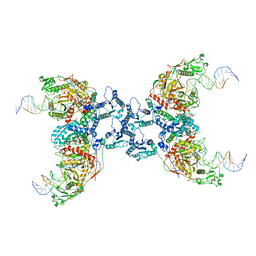 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA tetramer (NADase active form) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8K9G
 
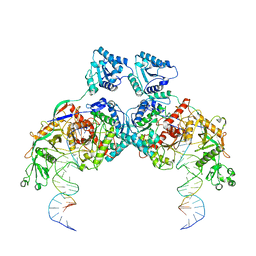 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-1) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
5KWB
 
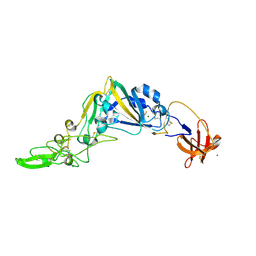 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 1.9 angstrom, molecular replacement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Spike glycoprotein, ... | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-17 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
5Y6N
 
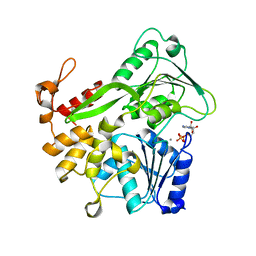 | | Zika virus helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase domain from Genome polyprotein, MANGANESE (II) ION | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
5Y6M
 
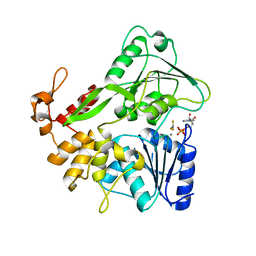 | | Zika virus helicase in complex with ADP-AlF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase domain from Genome polyprotein, ... | | Authors: | Yang, X.Y, Chen, C, Tian, H.L, Chi, H, Mu, Z.Y, Zhang, T.Q, Yang, K.L, Zhao, Q, Liu, X.H, Wang, Z.F, Ji, X.Y, Yang, H.T. | | Deposit date: | 2017-08-12 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Mechanism of ATP hydrolysis by the Zika virus helicase.
FASEB J., 32, 2018
|
|
7V2Z
 
 | | ZIKV NS3helicase in complex with ssRNA and ATP-Mn2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Core protein, MANGANESE (II) ION, ... | | Authors: | Lin, M.M, Yang, H.T. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.10101676 Å) | | Cite: | Structural Basis of Zika Virus Helicase in RNA Unwinding and ATP Hydrolysis.
Acs Infect Dis., 8, 2022
|
|
5GNB
 
 | | Crystal Structure of the Receptor Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1 (HKU1 1A-CTD, 2.3 angstrom, native-SAD phasing) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Guan, H, Wojdyla, J.A, Wang, M, Cui, S. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the receptor binding domain of the spike glycoprotein of human betacoronavirus HKU1
Nat Commun, 8, 2017
|
|
5ZT1
 
 | | Structure of the bacterial pathogens ATPase with substrate ATP gamma S | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Gao, X.P, Mu, Z.X, Cui, S. | | Deposit date: | 2018-05-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.114 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri
Front Microbiol, 9, 2018
|
|
5YBI
 
 | | Structure of the bacterial pathogens ATPase with substrate AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ATP synthase SpaL/MxiB, ... | | Authors: | Mu, Z.X, Gao, X.P, Cui, S. | | Deposit date: | 2017-09-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Structural Insight Into Conformational Changes Induced by ATP Binding in a Type III Secretion-Associated ATPase FromShigella flexneri.
Front Microbiol, 9, 2018
|
|
4ZHS
 
 | |
4ZIC
 
 | |
6V8Z
 
 | |
6V8X
 
 | | VRC01 Bound BG505 F14 HIV-1 SOSIP Envelope Trimer Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2019-12-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Disruption of the HIV-1 Envelope allosteric network blocks CD4-induced rearrangements.
Nat Commun, 11, 2020
|
|
8EU8
 
 | | Cryo-EM structure of CH848 10.17DT DS-SOSIP-2P Env | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848 10.17DT SOSIP Envelope glycoprotein gp160 | | Authors: | Wrapp, D, Acharya, P, Haynes, B.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure-Based Stabilization of SOSIP Env Enhances Recombinant Ectodomain Durability and Yield.
J.Virol., 97, 2023
|
|
7BH8
 
 | | 3H4-Fab HLA-E-VL9 co-complex | | Descriptor: | 3H4 Fab heavy chain, 3H4 Fab light chain, Beta-2-microglobulin, ... | | Authors: | Walters, L.C, Rozbesky, D. | | Deposit date: | 2021-01-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mouse and human antibodies bind HLA-E-leader peptide complexes and enhance NK cell cytotoxicity.
Commun Biol, 5, 2022
|
|
7EA8
 
 | |
7EA5
 
 | |
