5F0E
 
 | | Murine endoplasmic reticulum alpha-glucosidase II | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-11-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5H9O
 
 | | Complex of Murine endoplasmic reticulum alpha-glucosidase II with D-Glucose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2015-12-29 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HJO
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with bound substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-alpha-D-arabino-hexopyranose-(1-3)-D-glucal, ACETATE ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-01-13 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HJR
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with bound covalent intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoro-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-01-13 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IEG
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.822 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IEF
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with N-butyl-1-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-1-BUTYL-2-(HYDROXYMETHYL)PIPERIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IEE
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with 1-deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IED
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5MHF
 
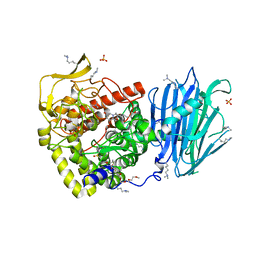 | | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mannosyl-oligosaccharide glucosidase, ... | | Authors: | Hill, J.C, Caputo, A.T, Roversi, P, Zitzmann, N. | | Deposit date: | 2016-11-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Endoplasmic Reticulum alpha-Glucosidase I with a Single-Dose Iminosugar Treatment Protects against Lethal Influenza and Dengue Virus Infections.
J.Med.Chem., 2020
|
|
7ZKC
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (apo form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Le Cornu, J.D. | | Deposit date: | 2022-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
7ZLU
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-2-deoxy-2-fluoro-D-glucose | | Descriptor: | 1,3-PROPANDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Ibba, R. | | Deposit date: | 2022-04-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
7ZLL
 
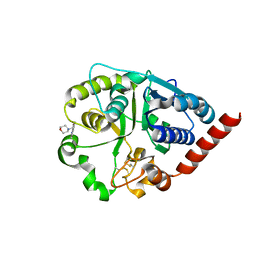 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with the 5-[(morpholin-4-yl)methyl]quinolin-8-ol inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(morpholin-4-ylmethyl)quinolin-8-ol, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Kantsadi, A.L, Chandran, A.V. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
7K9T
 
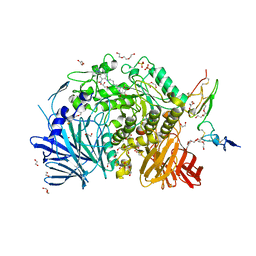 | | Co-crystal structure of alpha glucosidase with compound 5 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-{[(5Z)-6-{[2-nitro-4-(2H-1,2,3-triazol-2-yl)phenyl]amino}hex-5-en-1-yl]amino}cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9Q
 
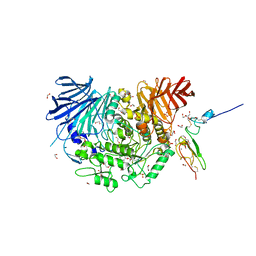 | | Co-crystal structure of alpha glucosidase with compound 4 | | Descriptor: | (1S,2S,3R,4S,5S)-5-amino-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9N
 
 | | Co-crystal structure of alpha glucosidase with compound 2 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(9-methoxynonyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7K9O
 
 | | Co-crystal structure of alpha glucosidase with compound 3 | | Descriptor: | (1~{S},2~{S},3~{R},4~{S},5~{S})-1-(hydroxymethyl)-5-[6-[[2-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]-4-(1,2,3,4-tetrazol-1-yl)phenyl]amino]hexylamino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Alpha glucosidase 2 alpha neutral subunit, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KBJ
 
 | | Co-crystal structure of alpha glucosidase with compound 9 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-{[2-(2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethoxy)ethyl]amino}cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KB6
 
 | | Co-crystal structure of alpha glucosidase with compound 7 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(pyrimidin-2-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KAD
 
 | | Co-crystal structure of alpha glucosidase with compound 6 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(6-{[2-nitro-4-(1H-1,2,3-triazol-1-yl)phenyl]amino}hexyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KB8
 
 | | Co-crystal structure of alpha glucosidase with compound 8 | | Descriptor: | (1S,2S,3R,4S,5S)-1-(hydroxymethyl)-5-[(4-{[2-nitro-4-(triazan-1-yl)phenyl]amino}butyl)amino]cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-01 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KBR
 
 | | Co-crystal structure of alpha glucosidase with compound 10 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7L9E
 
 | | Crystal structure of apo-alpha glucosidase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-01-03 | | Release date: | 2021-12-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7KRY
 
 | | Co-crystal structure of alpha glucosidase with compound 11 | | Descriptor: | (1S,2S,3R,4S,5S)-5-({6-[(4-azido-2-nitrophenyl)amino]hexyl}amino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | N-Substituted Valiolamine Derivatives as Potent Inhibitors of Endoplasmic Reticulum alpha-Glucosidases I and II with Antiviral Activity.
J.Med.Chem., 64, 2021
|
|
7GH2
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-090737b9-1 (Mpro-x12735) | | Descriptor: | (4R)-6-chloro-N-(isoquinolin-4-yl)-N-propanoyl-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GHE
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-916a2c5a-4 (Mpro-x3303) | | Descriptor: | 3C-like proteinase, 4-(4-phenylpiperazine-1-carbonyl)quinolin-2(1H)-one, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
