8HU2
 
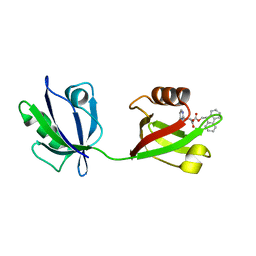 | | Rattus Syntenin-1 PDZ domain with inhibitor | | Descriptor: | (2~{S})-2-(9~{H}-fluoren-9-ylmethoxycarbonylamino)-3-(4-oxidanylidene-5~{H}-pyrimidin-2-yl)propanoic acid, Syntenin-1 | | Authors: | Heo, Y, Lee, J, Yun, J.H, Lee, W. | | Deposit date: | 2022-12-22 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of STNPDZ with inhibitor at 1.60 Angstroms resolution.
To Be Published
|
|
7X0D
 
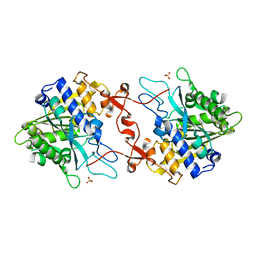 | | Crystal structure of phospholipase A1, CaPLA1 | | Descriptor: | Phospholipase A1, SULFATE ION | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39725161 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
7X0C
 
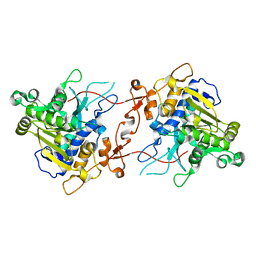 | | Crystal structure of phospholipase A1, AtDSEL | | Descriptor: | Phospholipase A1-IIgamma | | Authors: | Heo, Y, Lee, I, Moon, S, Lee, W. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79909515 Å) | | Cite: | Crystal Structures of the Plant Phospholipase A1 Proteins Reveal a Unique Dimerization Domain.
Molecules, 27, 2022
|
|
7DCK
 
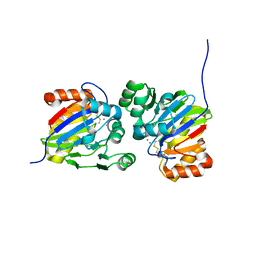 | | Crystal structure of phosphodiesterase tw9814 | | Descriptor: | Lactamase_B domain-containing protein, MANGANESE (II) ION | | Authors: | Heo, Y, Yun, J.H, Park, J.H, Park, S.B, Cha, S.S, Lee, W. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural and functional identification of the uncharacterized metallo-beta-lactamase superfamily protein TW9814 as a phosphodiesterase with unique metal coordination.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7CYU
 
 | | Crystal structure of human BAF57 HMG domain | | Descriptor: | SULFATE ION, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 | | Authors: | Heo, Y, Yun, J.H, Park, J.H, Lee, W. | | Deposit date: | 2020-09-04 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the HMG domain of human BAF57 and its interaction with four-way junction DNA.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7FDJ
 
 | | Engineered Hepatitis B virus core antigen with short linker T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-07-16 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EP6
 
 | | Engineered Hepatitis B virus core antigen T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EOY
 
 | | Engineered Hepatitis B virus core antigen T=3 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-24 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
8KHO
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in complex with two Cobalt ions and Methionine | | Descriptor: | COBALT (II) ION, METHIONINE, Methionine aminopeptidase 1D, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8KHN
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in complex with two cobalt ions | | Descriptor: | COBALT (II) ION, Methionine aminopeptidase 1D, mitochondrial, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8KHM
 
 | | Crystal structure of human methionine aminopeptidase 12 (MAP12) in the unbound form | | Descriptor: | GLYCEROL, Methionine aminopeptidase 1D, mitochondrial, ... | | Authors: | Lee, Y, Lee, E, Hahn, H, Kim, H, Heo, Y, Jang, D.M, Kim, H.J, Kim, H.S. | | Deposit date: | 2023-08-22 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural insights into N-terminal methionine cleavage by the human mitochondrial methionine aminopeptidase, MetAP1D.
Sci Rep, 13, 2023
|
|
8X9G
 
 | | Crystal structure of CO dehydrogenase mutant in complex with BV | | Descriptor: | 1-(phenylmethyl)-4-[1-(phenylmethyl)pyridin-1-ium-4-yl]pyridin-1-ium, Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9H
 
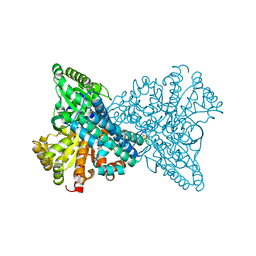 | | Crystal structure of CO dehydrogenase mutant (F41C) | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9F
 
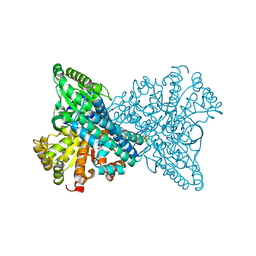 | | Crystal structure of CO dehydrogenase mutant in complex with EV | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-4-(1-ethylpyridin-1-ium-4-yl)pyridin-1-ium, Carbon monoxide dehydrogenase 2, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9E
 
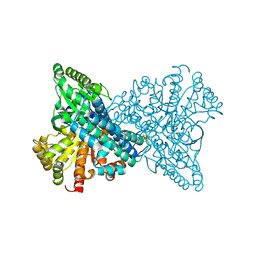 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in low PEG concentration | | Descriptor: | 1,2-ETHANEDIOL, Carbon monoxide dehydrogenase 2, FE (III) ION, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9D
 
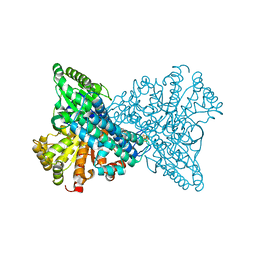 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in high PEG concentration | | Descriptor: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | Authors: | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | Deposit date: | 2023-11-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
7WJ5
 
 | | Cryo-EM structure of human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand binding specificity | | Descriptor: | Gai1 antibody (scfv16), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Heo, Y.S, Yoon, E.J, Jeon, Y.E, Yun, J.-H, Ishimoto, N, Woo, H, Park, S.Y, Song, J, Lee, W.T. | | Deposit date: | 2022-01-05 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM structure of the human somatostatin receptor 2 complex with its agonist somatostatin delineates the ligand-binding specificity.
Elife, 11, 2022
|
|
7XBD
 
 | | Cryo-EM structure of human galanin receptor 2 | | Descriptor: | Galanin, Galanin receptor type 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ishimoto, N, Kita, S, Park, S.Y. | | Deposit date: | 2022-03-21 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface.
Plos Biol., 20, 2022
|
|
6LST
 
 | | Crystal straucture of Uso1-1 | | Descriptor: | Intracellular protein transport protein USO1 | | Authors: | Heo, Y.Y, Lee, H.H. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of Uso1 membrane tether reveal an alternative conformation in the globular head domain
Sci Rep, 10, 2020
|
|
6LSU
 
 | | Crystal structure of Uso1-2 | | Descriptor: | Intracellular protein transport protein USO1 | | Authors: | Heo, Y.Y, Lee, H.H. | | Deposit date: | 2020-01-20 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Uso1 membrane tether reveal an alternative conformation in the globular head domain
Sci Rep, 10, 2020
|
|
7CFZ
 
 | | SH3 domain of NADPH oxidase activator 1 | | Descriptor: | NADPH oxidase activator 1 | | Authors: | Kim, M, Park, J.H, Attri, P, Lee, W. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural modification of NADPH oxidase activator (Noxa 1) by oxidative stress: An experimental and computational study.
Int.J.Biol.Macromol., 163, 2020
|
|
