8X94
 
 | | Structure of human TRPV1 in complex with antagonist --protein purified without CHS | | Descriptor: | 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile, CHOLESTEROL, Transient receptor potential cation channel subfamily V member 1,Green fluorescent protein | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-11-29 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Nat Commun, 15, 2024
|
|
2INF
 
 | | Crystal Structure of Uroporphyrinogen Decarboxylase from Bacillus subtilis | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Fan, J, Liu, Q, Hao, Q, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-10-06 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of uroporphyrinogen decarboxylase from Bacillus subtilis
J.Bacteriol., 189, 2007
|
|
8JQR
 
 | | Structure of human TRPV1 in complex with antagonist | | Descriptor: | 4-(7-Hydroxy-2-isopropyl-4-oxoquinazolin-3(4H)-yl)benzonitrile, CHOLESTEROL, Transient receptor potential cation channel subfamily V member 1,PreScission Site,Green fluorescent protein | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis of TRPV1 inhibition by SAF312 and cholesterol.
Nat Commun, 15, 2024
|
|
8JU6
 
 | | Structure of human TRPV4 with antagonist GSK279 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JVJ
 
 | | Structure of human TRPV4 with antagonist A2 and RhoA | | Descriptor: | Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JU5
 
 | | Structure of human TRPV4 with antagonist A1 | | Descriptor: | 4-[(3~{S},4~{S})-4-(aminomethyl)-1-(5-chloranylpyridin-2-yl)sulfonyl-4-oxidanyl-pyrrolidin-3-yl]oxy-2-fluoranyl-benzenecarbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JVI
 
 | | Structure of human TRPV4 with antagonist A2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
6V8Q
 
 | | Structure of an inner membrane protein required for PhoPQ regulated increases in outer membrane cardiolipin | | Descriptor: | (9Z,21R,24R,30R,33R,44Z)-24,27,30-trihydroxy-18,24,30,36-tetraoxo-19,23,25,29,31,35-hexaoxa-24lambda~5~,30lambda~5~-dip hosphatripentaconta-9,44-diene-21,33-diyl (9Z,9'Z)di-octadec-9-enoate, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fan, J, Miller, S. | | Deposit date: | 2019-12-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.699585 Å) | | Cite: | Structure of an Inner Membrane Protein Required for PhoPQ-Regulated Increases in Outer Membrane Cardiolipin.
Mbio, 11, 2020
|
|
4PX7
 
 | | Crystal structure of lipid phosphatase E. coli PgpB | | Descriptor: | GLYCEROL, LAURYL DIMETHYLAMINE-N-OXIDE, Phosphatidylglycerophosphatase | | Authors: | Fan, J, Jiang, D, Zhao, Y, Zhang, X.C. | | Deposit date: | 2014-03-22 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7DIX
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 5 | | Descriptor: | Na(+):neurotransmitter symporter (Snf family), SELENOMETHIONINE, SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJC
 
 | | Crystal structure of the G26C/Q250A mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ2
 
 | | Crystal structure of the G26C/E290S mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ1
 
 | | Crystal structure of the G26C mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.528 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DII
 
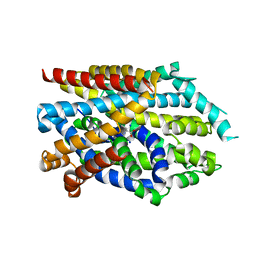 | | Crystal structure of LeuT in lipidic cubic phase at pH 7 | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7XJ2
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C4 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ1
 
 | | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ3
 
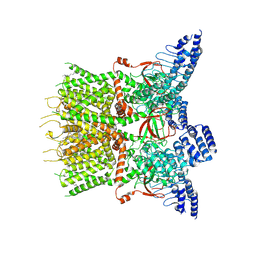 | | Structure of human TRPV3 | | Descriptor: | [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, fusion of transient receptor potential cation channel subfamily V member 3 and 3C-GFP | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7XJ0
 
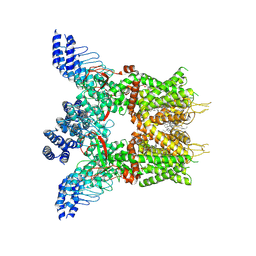 | | Structure of human TRPV3 in complex with Trpvicin | | Descriptor: | Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP, N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Fan, J, Yue, Z, Jiang, D, Lei, X. | | Deposit date: | 2022-04-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of TRPV3 inhibition by an antagonist.
Nat.Chem.Biol., 19, 2023
|
|
7UBU
 
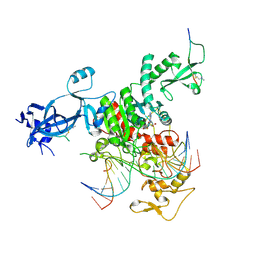 | |
7L4C
 
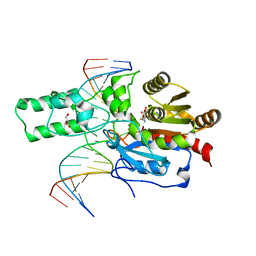 | | Crystal structure of the DRM2-CTT DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
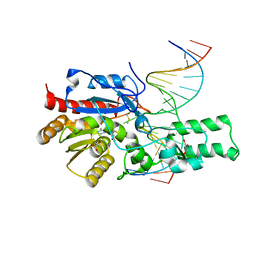 | | Crystal structure of the DRM2-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
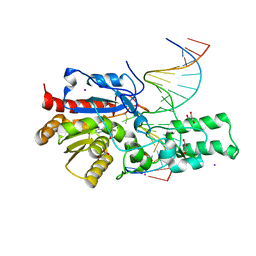 | | Crystal structure of the DRM2-CTG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
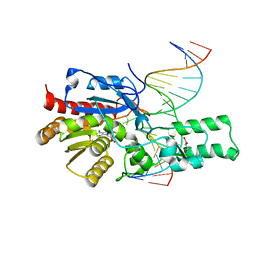 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
 | | Crystal structure of the DRM2-CCT DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
 | | Crystal structure of the DRM2-CAT DNA complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Fang, J, Song, J. | | Deposit date: | 2020-12-19 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
