8FFJ
 
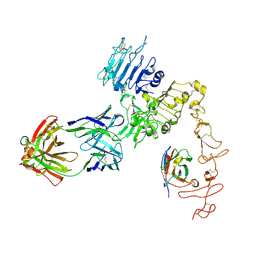 | | Structure of Zanidatamab bound to HER2 | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, Zanidatamab Heavy Chain A, Zanidatamab Heavy Chain B, ... | | Authors: | Worrall, L.J, Atkinson, C.E, Sanches, M, Dixit, S, Strynadka, N.C.J. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | An anti-HER2 biparatopic antibody that induces unique HER2 clustering and complement-dependent cytotoxicity.
Nat Commun, 14, 2023
|
|
7TTZ
 
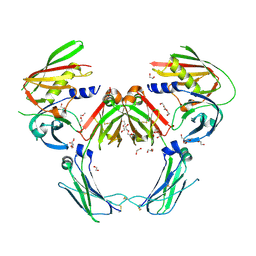 | | Heterodimeric IgA Fc in complex with Staphylococcus aureus protein SSL7 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Boulanger, M.J, Verstraete, M, Heinkel, F, Escobar, E, Dixit, S, Von Kreudenstein, T.S. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Engineering a pure and stable heterodimeric IgA for the development of multispecific therapeutics.
Mabs, 14, 2022
|
|
3UL3
 
 | |
1FWY
 
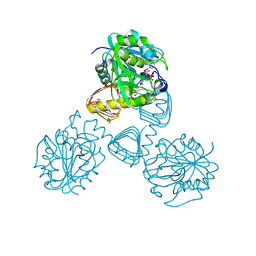 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE BOUND TO UDP-GLCNAC | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE, ... | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-25 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
1FXJ
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
6HKE
 
 | | MatC (Rpa3494) from Rhodopseudomonas palustris with bound malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, D-MALATE, Possible TctC subunit of the Tripartite Tricarboxylate Transport(TTT) Family | | Authors: | Rosa, L.T, Dix, S, Rafferty, J.B, Kelly, D.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | A New Mechanism for High-Affinity Uptake of C4-Dicarboxylates in Bacteria Revealed by the Structure of Rhodopseudomonas palustris MatC (RPA3494), a Periplasmic Binding Protein of the Tripartite Tricarboxylate Transporter (TTT) Family.
J. Mol. Biol., 431, 2019
|
|
7OGT
 
 | | Folded elbow of cohesin | | Descriptor: | Structural maintenance of chromosomes protein 1, Structural maintenance of chromosomes protein 3 | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Patele, N.J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Elife, 10, 2021
|
|
8D36
 
 | |
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8D6Z
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV91-27 | | Descriptor: | Neutralizing antibody COV91-27 heavy chain, Neutralizing antibody COV91-27 light chain, Spike protein S2 fusion peptide | | Authors: | Lee, C.C.D, Lin, T.H, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
8DTX
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV89-22 | | Descriptor: | COV89-22 heavy chain, COV89-22 light chain, Spike protein S2' stem helix peptide | | Authors: | Lin, T.H, Lee, C.C.D, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTR
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV30-14 | | Descriptor: | COV30-14 heavy chain, COV30-14 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
8DTT
 
 | | Crystal structure of SARS-CoV-2 spike stem helix peptide in complex with neutralizing antibody COV93-03 | | Descriptor: | COV93-03 heavy chain, COV93-03 light chain, Spike protein S2' stem helix peptide | | Authors: | Lee, C.C.D, Lin, T.H, Wilson, I.A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rare, convergent antibodies targeting the stem helix broadly neutralize diverse betacoronaviruses.
Cell Host Microbe, 31, 2023
|
|
7DG5
 
 | |
3PLZ
 
 | | Human LRH1 LBD bound to GR470 | | Descriptor: | (3aS,6aR)-5-[(4E)-oct-4-en-4-yl]-N,4-diphenyl-2,3,6,6a-tetrahydropentalen-3a(1H)-amine, 1,2-ETHANEDIOL, FTZ-F1 related protein, ... | | Authors: | Williams, S.P, Xu, R, Zuercher, W.J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Agonists of the Orphan Nuclear Receptors Steroidogenic Factor-1 (SF-1, NR5A1) and Liver Receptor Homologue-1 (LRH-1, NR5A2).
J.Med.Chem., 54, 2011
|
|
