1FJ1
 
 | |
9L99
 
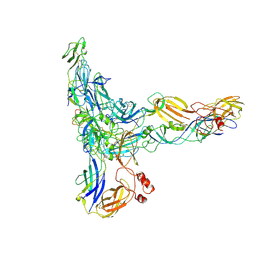 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA1-2 | | Descriptor: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | Deposit date: | 2024-12-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
9L9A
 
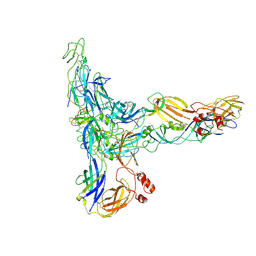 | | Structure of Western equine encephalitis virus McMillan strain in complex with VLDLR LA2-3 | | Descriptor: | CALCIUM ION, Spike glycoprotein E1, Spike glycoprotein E2, ... | | Authors: | Ma, B, Cao, Z, Ding, W, Zhang, X, Xiang, Y, Cao, D. | | Deposit date: | 2024-12-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for the recognition of two different types of receptors by Western equine encephalitis virus.
Cell Rep, 44, 2025
|
|
8HN9
 
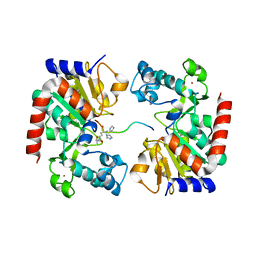 | | Human SIRT3 Recognizing CCNE2K348la peptide | | Descriptor: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Wang, Y, Ding, W. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
8IWZ
 
 | | Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
4NOK
 
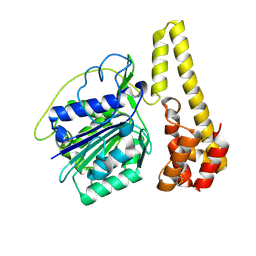 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOM
 
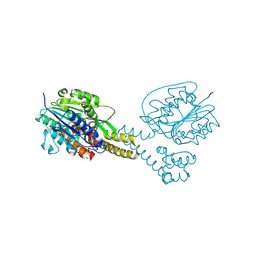 | | Crystal structure of asparaginyl endopeptidase (AEP)/Legumain activated at pH 4.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOJ
 
 | | Crystal structure of the mature form of asparaginyl endopeptidase (AEP)/Legumain activated at pH 3.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
4NOL
 
 | | Crystal structure of proenzyme asparaginyl endopeptidase (AEP)/Legumain mutant D233A at pH 7.5 | | Descriptor: | Legumain | | Authors: | Zhao, L, Hua, T, Ru, H, Ni, X, Shaw, N, Jiao, L, Ding, W, Qu, L, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-11-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of asparaginyl endopeptidase reveals the activation mechanism and a reversible intermediate maturation stage.
Cell Res., 24, 2014
|
|
8J4I
 
 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
8IWX
 
 | | Cryo-EM structure of unprotonated LHCII in detergent solution at high pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IWY
 
 | | Cryo-EM structure of protonated LHCII in detergent solution at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IX1
 
 | | Cryo-EM structure of protonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
4M4E
 
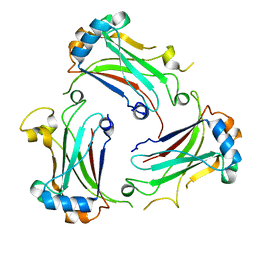 | | TRAF domain of human TRAF4 | | Descriptor: | TNF receptor-associated factor 4 | | Authors: | Niu, F, Ru, H, Ding, W, Ouyang, S, Liu, Z.J. | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural biology study of human TNF receptor associated factor 4 TRAF domain
Protein Cell, 4, 2013
|
|
8IT0
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-2) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISY
 
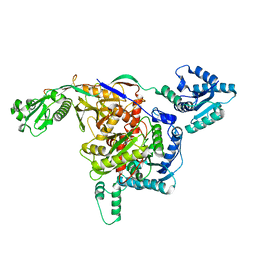 | | Cryo-EM structure of free-state Crt-SPARTA | | Descriptor: | Piwi domain-containing protein, TIR domain-containing protein | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8ISZ
 
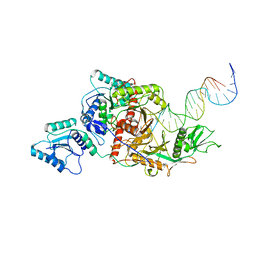 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA monomer | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IT1
 
 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA tetramer (NADase active form) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-03-21 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
4MRX
 
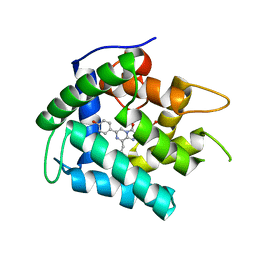 | | Crystal Structure of Y138F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8K9G
 
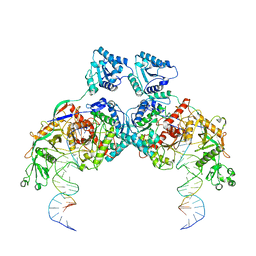 | | Cryo-EM structure of Crt-SPARTA-gRNA-tDNA dimer (conformation-1) | | Descriptor: | DNA (45-mer), Piwi domain-containing protein, RNA (5'-R(P*UP*GP*AP*GP*GP*UP*AP*GP*UP*AP*GP*GP*UP*UP*GP*UP*AP*UP*AP*GP*U)-3'), ... | | Authors: | Gao, X, Shang, K, Zhu, K, Wang, L, Mu, Z, Fu, X, Yu, X, Qin, B, Zhu, H, Ding, W, Cui, S. | | Deposit date: | 2023-08-01 | | Release date: | 2023-10-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Nucleic-acid-triggered NADase activation of a short prokaryotic Argonaute.
Nature, 625, 2024
|
|
8IX0
 
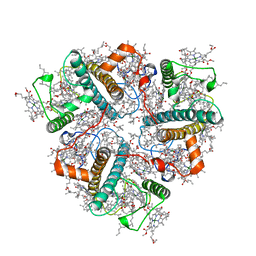 | | Cryo-EM structure of unprotonated LHCII nanodisc at high pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
8IX2
 
 | | Cryo-EM structure of unprotonated LHCII nanodisc at low pH value | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Ruan, M.X, Ding, W. | | Deposit date: | 2023-03-31 | | Release date: | 2023-09-06 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation.
Nat.Plants, 9, 2023
|
|
4MRY
 
 | | Crystal Structure of Ca(2+)- discharged Y138F obelin mutant from Obelia longissima at 1.30 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4QGU
 
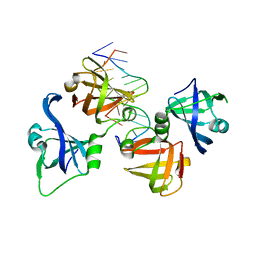 | | protein domain complex with ssDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*CP*CP*GP*GP*CP*GP*TP*GP*A)-3'), Gamma-interferon-inducible protein 16 | | Authors: | Ni, X, Ru, H, Zhao, L, Shaw, N, Ding, W, Songying, O, Liu, Z.-J. | | Deposit date: | 2014-05-25 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | New insights into the structural basis of DNA recognition by HINa and HINb domains of IFI16.
J Mol Cell Biol, 8, 2016
|
|
4PW8
 
 | | Human tryptophan 2,3-dioxygenase | | Descriptor: | COBALT (II) ION, Tryptophan 2,3-dioxygenase | | Authors: | Meng, B, Wu, D, Gu, J.H, Ouyang, S.Y, Ding, W, Liu, Z.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and functional analyses of human tryptophan 2,3-dioxygenase
Proteins, 82, 2014
|
|
