1NTP
 
 | |
1TGN
 
 | |
3PGF
 
 | | Crystal structure of maltose bound MBP with a conformationally specific synthetic antigen binder (sAB) | | Descriptor: | GLYCEROL, IMIDAZOLE, Maltose-binding periplasmic protein, ... | | Authors: | Kossiakoff, A.A, Duguid, E.M, Sandstrom, A. | | Deposit date: | 2010-11-01 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allosteric control of ligand-binding affinity using engineered conformation-specific effector proteins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1D9C
 
 | | BOVINE INTERFERON-GAMMA AT 2.0 ANGSTROMS | | Descriptor: | INTERFERON-GAMMA | | Authors: | Randal, M, Kossiakoff, A.A. | | Deposit date: | 1999-10-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of bovine interferon-gamma; assessment of the structural differences between species.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1D9G
 
 | | BOVINE INTERFERON-GAMMA AT 2.9 ANGSTROMS | | Descriptor: | INTERFERON-GAMMA | | Authors: | Randal, M, Kossiakoff, A.A. | | Deposit date: | 1999-10-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.0 A structure of bovine interferon-gamma; assessment of the structural differences between species.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3HHR
 
 | |
5W2B
 
 | | Crystal structure of C-terminal domain of Ebola (Reston) nucleoprotein in complex with Fab fragment | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Nucleoprotein | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of the C-terminal domain of the nucleoprotein from the Bundibugyo strain of the Ebola virus in complex with a pan-specific synthetic Fab.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5EII
 
 | |
8PG0
 
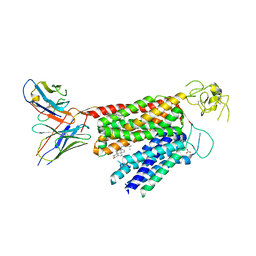 | | Human OATP1B3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHOLESTEROL, ... | | Authors: | Ciuta, A.-D, Nosol, K, Kowal, J, Mukherjee, S, Ramirez, A.S, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2023-06-17 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of human drug transporters OATP1B1 and OATP1B3.
Nat Commun, 14, 2023
|
|
8PHW
 
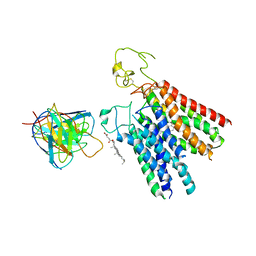 | | Human OATP1B1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Fab18 (heavy chain, variable region), ... | | Authors: | Ciuta, A.-D, Nosol, K, Kowal, J, Mukherjee, S, Ramirez, A.S, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2023-06-20 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of human drug transporters OATP1B1 and OATP1B3.
Nat Commun, 14, 2023
|
|
5UEA
 
 | |
5UEK
 
 | |
7ZYI
 
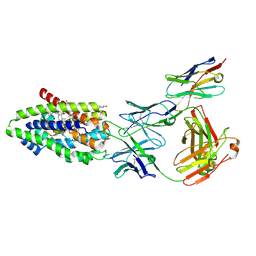 | | Structure of the human sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody | | Descriptor: | CHOLESTEROL, GLYCOCHENODEOXYCHOLIC ACID, Nanobody, ... | | Authors: | Liu, H, Irobalieva, R.N, Bang-Sorensen, R, Nosol, K, Mukherjee, S, Agrawal, P, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver.
Cell Res., 32, 2022
|
|
7ZLH
 
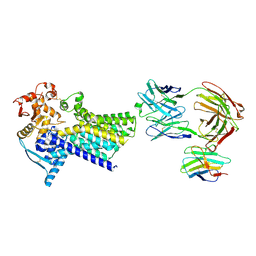 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
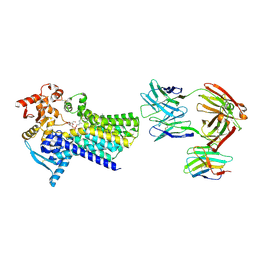 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLG
 
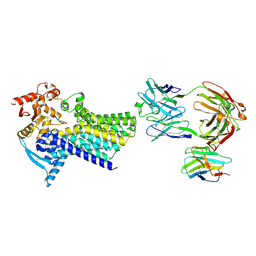 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Mao, R, Irobalieva, R, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLJ
 
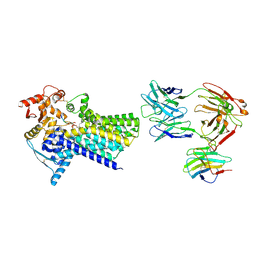 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
4XTR
 
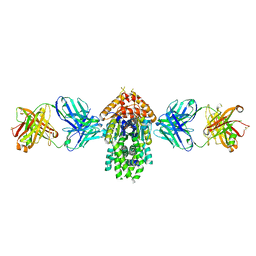 | | Structure of Get3 bound to the transmembrane domain of Pep12 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XVU
 
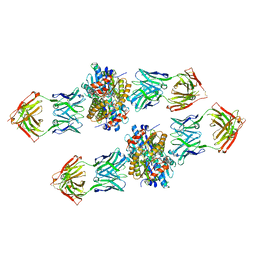 | | Structure of Get3 bound to the transmembrane domain of Nyv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, Antibody heavy chain, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XH2
 
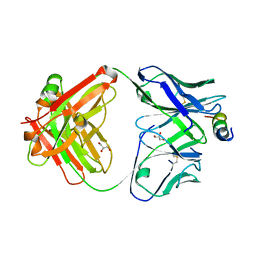 | | Crystal structure of human paxillin LD4 motif in complex with Fab fragment | | Descriptor: | ACETATE ION, ACETYL GROUP, Fab Heavy Chain, ... | | Authors: | Nocula-Lugowska, M, Lugowski, M, Salgia, R, Kossiakoff, A.A. | | Deposit date: | 2015-01-04 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering Synthetic Antibody Inhibitors Specific for LD2 or LD4 Motifs of Paxillin.
J.Mol.Biol., 427, 2015
|
|
1F6F
 
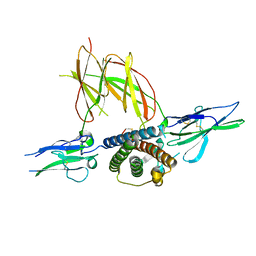 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX BETWEEN OVINE PLACENTAL LACTOGEN AND THE EXTRACELLULAR DOMAIN OF THE RAT PROLACTIN RECEPTOR | | Descriptor: | PLACENTAL LACTOGEN, PROLACTIN RECEPTOR | | Authors: | Elkins, P.A, Christinger, H.W, Sandowski, Y, Sakal, E, Gertler, A, De Vos, A.M, Kossiakoff, A.A. | | Deposit date: | 2000-06-21 | | Release date: | 2000-07-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor.
Nat.Struct.Biol., 7, 2000
|
|
4XMN
 
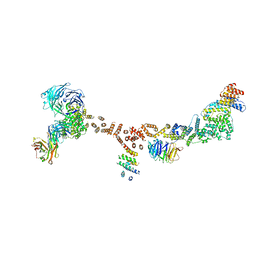 | | Structure of the yeast coat nucleoporin complex, space group P212121 | | Descriptor: | Antibody 87 heavy chain, Antibody 87 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XWO
 
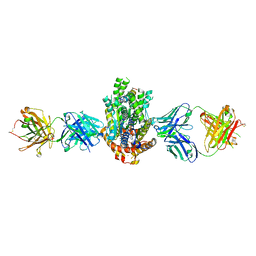 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
4XMM
 
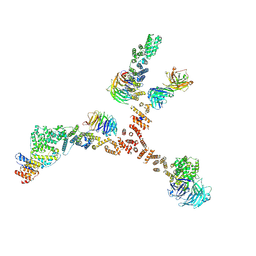 | | Structure of the yeast coat nucleoporin complex, space group C2 | | Descriptor: | Antibody 57 heavy chain, Antibody 57 light chain, Nucleoporin NUP120, ... | | Authors: | Stuwe, T, Correia, A.R, Lin, D.H, Paduch, M, Lu, V.T, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7.384 Å) | | Cite: | Nuclear pores. Architecture of the nuclear pore complex coat.
Science, 347, 2015
|
|
4XGZ
 
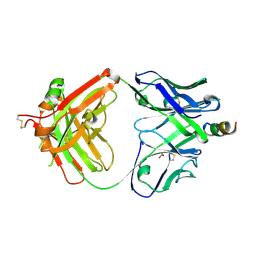 | | Crystal structure of human paxillin LD2 motif in complex with Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, FAB HEAVY CHAIN, FAB LIGHT CHAIN, ... | | Authors: | Nocula-Lugowska, M, Lugowski, M, Salgia, R, Kossiakoff, A.A. | | Deposit date: | 2015-01-04 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineering Synthetic Antibody Inhibitors Specific for LD2 or LD4 Motifs of Paxillin.
J.Mol.Biol., 427, 2015
|
|
