6FQB
 
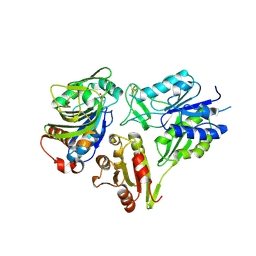 | | MurT/GatD peptidoglycan amidotransferase complex from Streptococcus pneumoniae R6 | | Descriptor: | Cobyric acid synthase, GLUTAMINE, Mur ligase family protein | | Authors: | Morlot, C, Contreras-Martel, C, Leisico, F, Straume, D, Peters, K, Hegnar, O.A, Simon, N, Villard, A.M, Breukink, E, Gravier-Pelletier, C, Le Corre, L, Vollmer, W, Pietrancosta, N, Havarstein, L.S, Zapun, A. | | Deposit date: | 2018-02-13 | | Release date: | 2018-08-22 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
6G1Y
 
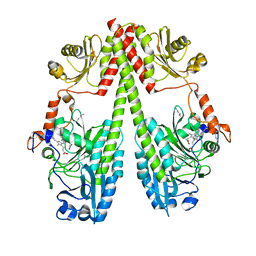 | | Crystal structure of the photosensory core module (PCM) of a bathy phytochrome from Agrobacterium fabrum in the Pfr state. | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Schmidt, A, Qureshi, B.M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
6G2H
 
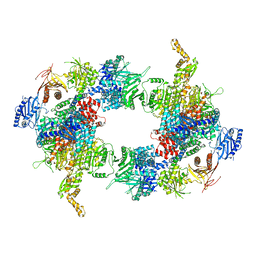 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6G73
 
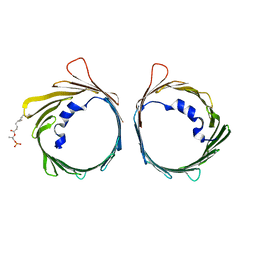 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 3.3 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
6G9Q
 
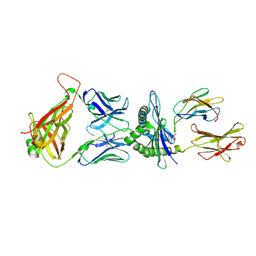 | |
6GCU
 
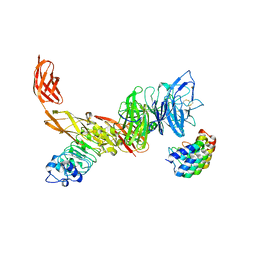 | | MET receptor in complex with InlB internalin domain and DARPin A3A | | Descriptor: | DARPin A3A, Hepatocyte growth factor receptor, Internalin B | | Authors: | Meyer, T, Andres, F, Iamele, L, Gherardi, E, Pluckthun, A, Niemann, H.H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.001 Å) | | Cite: | Inhibition of the MET Kinase Activity and Cell Growth in MET-Addicted Cancer Cells by Bi-Paratopic Linking.
J.Mol.Biol., 431, 2019
|
|
6G9Z
 
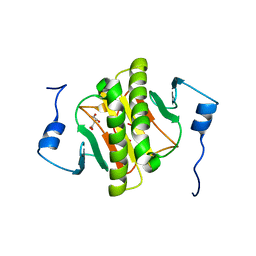 | |
6FP9
 
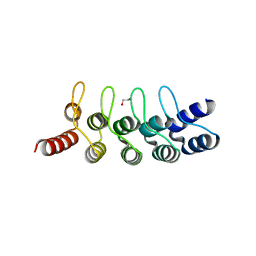 | | Crystal structure of anti-mTFP1 DARPin 1238_E11 | | Descriptor: | 1,2-ETHANEDIOL, DARPin 1238_E11, SULFATE ION | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6GBO
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Ebola virus | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
6FMK
 
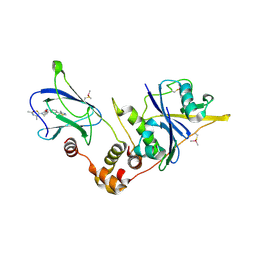 | | pVHL:EloB:EloC in complex with N-((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamothioyl) pyrrolidin-1-yl)-1-thioxopropan-2-yl)acetamide (ligand 4) | | Descriptor: | Elongin-B, Elongin-C, von Hippel-Lindau disease tumor suppressor, ... | | Authors: | Soares, P, Lucas, X, Ciulli, A. | | Deposit date: | 2018-01-31 | | Release date: | 2018-04-11 | | Last modified: | 2018-06-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Thioamide substitution to probe the hydroxyproline recognition of VHL ligands.
Bioorg. Med. Chem., 26, 2018
|
|
6FQT
 
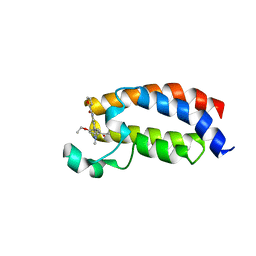 | | Crystal structure of CREBBP bromodomain complexd with DR46 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexd with DR46
To Be Published
|
|
6FQV
 
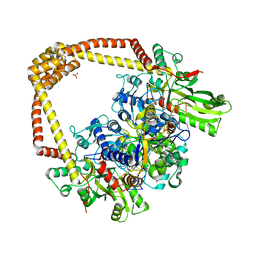 | | 2.60A BINARY COMPLEX OF S.AUREUS GYRASE with UNCLEAVED DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, DNA gyrase subunit B,DNA gyrase subunit B, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
6FSE
 
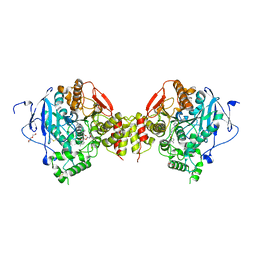 | | Mus musculus acetylcholinesterase in complex with 1-(4-(4-Ethylpiperazin-1-yl)piperidin-1-yl)-2-((4'-methoxy-[1,1'-biphenyl]-4-yl)oxy)ethanone dihydrochloride (15) | | Descriptor: | 1-[4-(4-ethylpiperazin-1-yl)piperidin-1-yl]-2-[4-(4-methoxyphenyl)phenoxy]ethanone, 2-(2-METHOXYETHOXY)ETHANOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ... | | Authors: | Knutsson, S, Engdahl, C, Kumari, R, Kindahl, T, Forsgren, N, Lindgren, C, Kitur, S, Kamau, L, Ekstrom, F, Linusson, A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent Inhibitors of Mosquito Acetylcholinesterase 1 with Resistance-Breaking Potency.
J.Med.Chem., 61, 2018
|
|
6FRK
 
 | | Structure of a prehandover mammalian ribosomal SRP and SRP receptor targeting complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Kobayashi, K, Jomaa, A, Ban, N. | | Deposit date: | 2018-02-16 | | Release date: | 2018-03-28 | | Last modified: | 2018-05-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a prehandover mammalian ribosomal SRP·SRP receptor targeting complex.
Science, 360, 2018
|
|
6FZ9
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2463 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FVU
 
 | | 26S proteasome, s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
8CYS
 
 | | CryoEM structures of amplified alpha-synuclein fibril class B type II with extended core from DLB case I | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYV
 
 | | CryoEM structures of amplified alpha-synuclein fibril class B mixed type I/II with extended core from DLB case II | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ2
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CZ6
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with extended core from DLB case X | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYX
 
 | | CryoEM structure of amplified alpha-synuclein fibril class A type I with compact core from DLB case III | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYY
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B mixed type I/II with extended core from DLB case V | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
6NQW
 
 | | Flagellar protein FcpA from Leptospira biflexa - hexagonal form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Flagellar coiling protein A, GLYCEROL, ... | | Authors: | San Martin, F, Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete
Elife, 9, 2020
|
|
8CZ0
 
 | | CryoEM structure of amplified alpha-synuclein fibril class B type I with extended core from DLB case VII | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
8CYW
 
 | | CryoEM structures of amplified alpha-synuclein fibril class B type I with compact core from DLB case III | | Descriptor: | Alpha-synuclein | | Authors: | Zhou, Y, Sokratian, A, Xu, E, Viverette, E, Dillard, L, Yuan, Y, Li, J.Y, Matarangas, A, Bouvette, J, Borgnia, M, Bartesaghi, A, West, A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional landscape of alpha-synuclein fibril assemblies amplified from cerebrospinal fluid
To Be Published
|
|
