4K5L
 
 | |
4K5O
 
 | |
4BTV
 
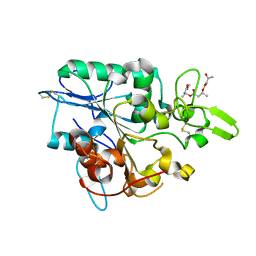 | | Structure of PhaZ7 PHB depolymerase in complex with 3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, PHB DEPOLYMERASE PHAZ7 | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4EME
 
 | |
2MAU
 
 | |
2MUR
 
 | |
6J66
 
 | | Chondroitin sulfate/dermatan sulfate endolytic 4-O-sulfatase | | Descriptor: | CALCIUM ION, Chondroitin sulfate/dermatan sulfate 4-O-endosulfatase protein | | Authors: | Gu, L, Li, F, Su, T, Wang, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases With High Identity.
Front Microbiol, 10, 2019
|
|
5WIV
 
 | | Structure of the sodium-bound human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
5WIU
 
 | | Structure of the human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
2DUT
 
 | |
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7YH5
 
 | | MazG(Mycobacterium tuberculosis) | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate pyrophosphohydrolase | | Authors: | Li, J, Wang, S. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of the housecleaning nucleoside triphosphate pyrophosphohydrolase MazG from Mycobacterium tuberculosis.
Front Microbiol, 14, 2023
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8JT6
 
 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
5AM6
 
 | | Native FGFR1 with an inhibitor | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Bunney, T.D, Wan, S, Thiyagarajan, N, Sutto, L, Williams, S.V, Ashford, P, Koss, H, Knowles, M.A, Gervasio, F.L, Coveney, P.V, Katan, M. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
5AM7
 
 | | FGFR1 mutant with an inhibitor | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Bunney, T.D, Wan, S, Thiyagarajan, N, Sutto, L, Williams, S.V, Ashford, P, Koss, H, Knowles, M.A, Gervasio, F.L, Coveney, P.V, Katan, M. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
3CVS
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(8OG)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CWA
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*(8OG)P*DAP*DCP*DAP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3D4V
 
 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
2GGT
 
 | | Crystal structure of human SCO1 complexed with nickel. | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, SCO1 protein homolog, ... | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Martinelli, M, Palumaa, P, Wang, S. | | Deposit date: | 2006-03-24 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3CM1
 
 | |
2RLI
 
 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
