8EO0
 
 | |
8ENW
 
 | |
6V6G
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V6P
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V8V
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-2 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-12 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V83
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-1 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V5E
 
 | | Crystal structure of CTX-M-14 P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6V7T
 
 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase in complex with ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
2IIT
 
 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T, Weber, A.E. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7KXJ
 
 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 3-"up", asymmetric | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
2IIV
 
 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-METHYL-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Weber, A.E, Biftu, T. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7KXK
 
 | | SARS-CoV-2 spike protein in complex with Fab 15033-7, 2-"up"-1-"down" conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 15033-7 heavy chain, ... | | Authors: | Li, Z, Rini, J. | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Tetravalent SARS-CoV-2 Neutralizing Antibodies Show Enhanced Potency and Resistance to Escape Mutations.
J.Mol.Biol., 433, 2021
|
|
1IVG
 
 | | STRUCTURES OF AROMATIC INHIBITORS OF INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, INFLUENZA A SUBTYPE N2 NEURAMINIDASE, ... | | Authors: | Jedrzejas, M.J, Luo, M. | | Deposit date: | 1994-12-12 | | Release date: | 1995-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of aromatic inhibitors of influenza virus neuraminidase.
Biochemistry, 34, 1995
|
|
8DZ8
 
 | |
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
6LFH
 
 | |
6LYM
 
 | | Crystal structure of D657A mutant of formylglycinamidine synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYO
 
 | | Crystal Structure of H296A mutant of Formylglycinamidine Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-15 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYK
 
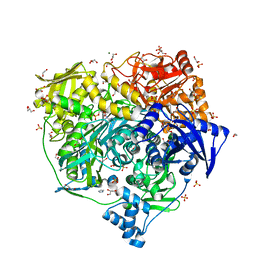 | | Crystal Structure of R1263A mutant of Formylglycinamidine Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYL
 
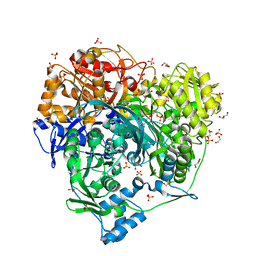 | | Crystal structure of S1052D mutant of Formylglycinamidine synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
2XCI
 
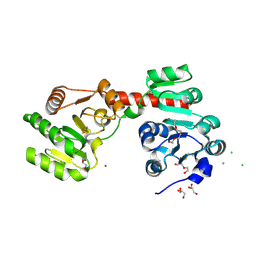 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XCU
 
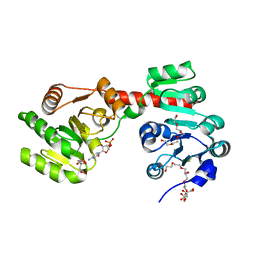 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5TVS
 
 | | JMJD2A in complex with Ni(II) | | Descriptor: | Lysine-specific demethylase 4A, NICKEL (II) ION, ZINC ION | | Authors: | Cascella, B, Lee, S.G, Jez, J.M. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | The small molecule JIB-04 disrupts O2 binding in the Fe-dependent histone demethylase KDM4A/JMJD2A.
Chem. Commun. (Camb.), 53, 2017
|
|
5TVR
 
 | | JMJD2A in complex with Ni(II) and alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Cascella, B, Lee, S.G, Jez, J.M. | | Deposit date: | 2016-11-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | The small molecule JIB-04 disrupts O2 binding in the Fe-dependent histone demethylase KDM4A/JMJD2A.
Chem. Commun. (Camb.), 53, 2017
|
|
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
