4TSV
 
 | |
3R7G
 
 | | Crystal structure of Spire KIND domain in complex with the tail of FMN2 | | Descriptor: | Formin-2, Protein spire homolog 1 | | Authors: | Kreutz, B, Vizcarra, C.L, Rodal, A.A, Toms, A.V, Lu, J, Quinlan, M.E, Eck, M.J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Spire KIND domain and insights into its interaction with Fmn-family formins
Proc.Natl.Acad.Sci.USA, 2011
|
|
3RBW
 
 | | Crystal structure of Spire KIND domain | | Descriptor: | Protein spire homolog 1 | | Authors: | Vizcarra, C.L, Kreutz, B, Rodal, A.A, Toms, A.V, Lu, J, Zheng, W, Quinlan, M.E, Eck, M.J. | | Deposit date: | 2011-03-30 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Spire KIND domain and insights into its interaction with Fmn-family formins
Proc.Natl.Acad.Sci.USA, 2011
|
|
7JVX
 
 | | Crystal structure of PTEN (aa 7-353 followed by spacer TGGGSGGTGGGSGGTGGGCY ligated to peptide pSDpTpTDpSDPENEPFDED) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JUL
 
 | | Crystal structure of non phosphorylated PTEN (n-crPTEN-13sp-T1, SDTTDSDPENEG) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-20 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7JTX
 
 | |
7JUK
 
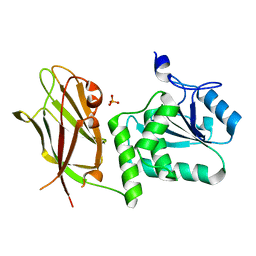 | | Crystal structure of PTEN with a tetra-phosphorylated tail (4p-crPTEN-13sp-T2, SDTTDSDPENEG) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-19 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1EXT
 
 | |
2TUN
 
 | |
1Y64
 
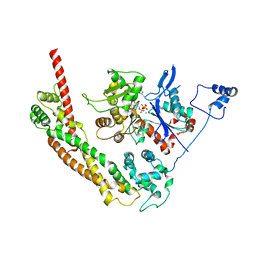 | | Bni1p Formin Homology 2 Domain complexed with ATP-actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Otomo, T, Tomchick, D.R, Otomo, C, Panchal, S.C, Machius, M, Rosen, M.K. | | Deposit date: | 2004-12-03 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain
Nature, 433, 2005
|
|
5TSW
 
 | |
1TCE
 
 | | SOLUTION NMR STRUCTURE OF THE SHC SH2 DOMAIN COMPLEXED WITH A TYROSINE-PHOSPHORYLATED PEPTIDE FROM THE T-CELL RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PHOSPHOPEPTIDE OF THE ZETA CHAIN OF T CELL RECEPTOR, SHC | | Authors: | Zhou, M.-M, Meadows, R.P, Logan, T.M, Yoon, H.S, Wade, W.R, Ravichandran, K.S, Burakoff, S.J, Feisk, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Shc SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1NCF
 
 | |
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
8D2J
 
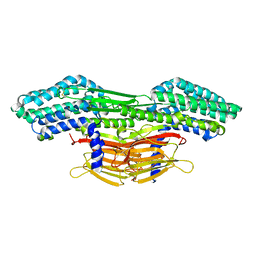 | | A novel insecticidal protein from ferns IPD113_Cow. | | Descriptor: | GLYCEROL, IPD113_Cow | | Authors: | Maher, M.J. | | Deposit date: | 2022-05-30 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel insecticidal proteins from ferns resemble insecticidal proteins from Bacillus thuringiensis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6G76
 
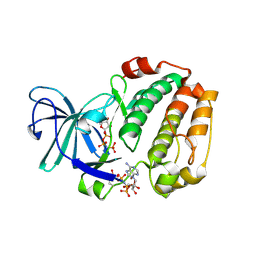 | |
6G78
 
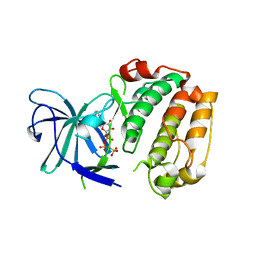 | | RSK4 N-terminal Kinase Domain S232E in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-6 | | Authors: | Prischi, F, Ali, M.M. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer.
Sci Transl Med, 13, 2021
|
|
6G77
 
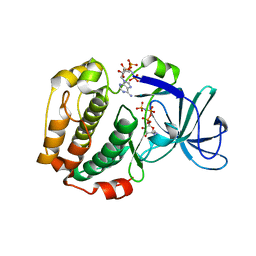 | | RSK4 N-terminal Kinase Domain in complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-6, ZINC ION | | Authors: | Prischi, F, Ali, M.M. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Repurposed floxacins targeting RSK4 prevent chemoresistance and metastasis in lung and bladder cancer.
Sci Transl Med, 13, 2021
|
|
7U0L
 
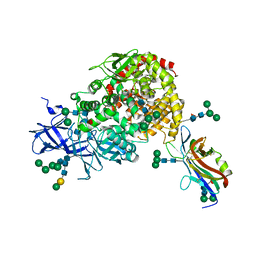 | | Crystal structure of the CCoV-HuPn-2018 RBD (domain B) in complex with canine APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USB
 
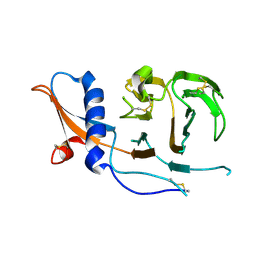 | | CCoV-HuPn-2018 S in the swung out conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US9
 
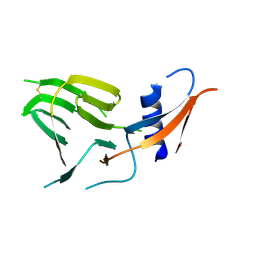 | | CCoV-HuPn-2018 S in the proximal conformation (local refinement of domain 0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7US6
 
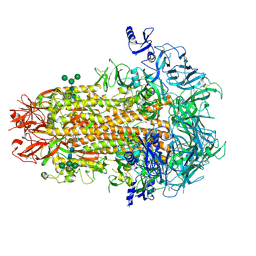 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the proximal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
7USA
 
 | | Structure of the human coronavirus CCoV-HuPn-2018 spike glycoprotein with domain 0 in the swung out conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Tortorici, M.A, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-04-23 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, receptor recognition, and antigenicity of the human coronavirus CCoV-HuPn-2018 spike glycoprotein.
Cell, 185, 2022
|
|
3I08
 
 | |
