1MTW
 
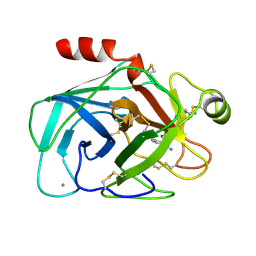 | | FACTOR XA SPECIFIC INHIBITOR IN COMPLEX WITH BOVINE TRYPSIN | | Descriptor: | (2S)-3-(7-carbamimidoylnaphthalen-2-yl)-2-[4-({(3R)-1-[(1Z)-ethanimidoyl]pyrrolidin-3-yl}oxy)phenyl]propanoic acid, CALCIUM ION, TRYPSIN | | Authors: | Stubbs, M.T. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of factor Xa specific inhibitors in complex with trypsin: structural grounds for inhibition of factor Xa and selectivity against thrombin.
FEBS Lett., 375, 1995
|
|
3QT5
 
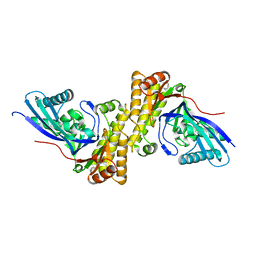 | | Crystal structure of Staphylococcus epidermidis mevalonate diphosphate decarboxylase | | Descriptor: | Mevalonate diphosphate decarboxylase | | Authors: | Barta, M.L, Skaff, A.D, McWhorter, W.J, Miziorko, H.M, Geisbrecht, B.V. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structures of Staphylococcus epidermidis mevalonate diphosphate decarboxylase bound to inhibitory analogs reveal new insight into substrate binding and catalysis.
J.Biol.Chem., 286, 2011
|
|
3QUI
 
 | | Crystal structure of Pseudomonas aeruginosa Hfq in complex with ADPNP | | Descriptor: | ADENINE, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Murina, V.N, Gabdulkhakov, A.G. | | Deposit date: | 2011-02-24 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1ADU
 
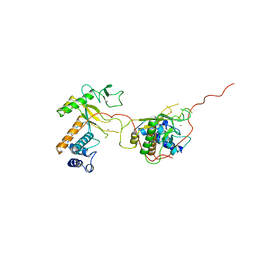 | | EARLY E2A DNA-BINDING PROTEIN | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, ZINC ION | | Authors: | Tucker, P.A, Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C. | | Deposit date: | 1995-05-11 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Alternative arrangements of the protein chain are possible for the adenovirus single-stranded DNA binding protein.
J.Mol.Biol., 257, 1996
|
|
1AJ6
 
 | |
4GRB
 
 | | Casein kinase 2 (CK2) bound to inhibitor | | Descriptor: | 5-(2-{[4-(dimethylcarbamoyl)phenyl]amino}-4-methoxypyrimidin-5-yl)thiophene-3-carboxylic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J.E, Ferguson, A.D. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo.
ACS Med Chem Lett, 4, 2013
|
|
4YG2
 
 | | X-ray crystal structur of Escherichia coli RNA polymerase sigma70 holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme
J. Biol. Chem., 288, 2013
|
|
1PHO
 
 | |
4Y7B
 
 | | Factor Xa complex with GTC000441 | | Descriptor: | 6-chloro-N-{(3S)-1-[(2S)-1-{(1R,5S)-7-[2-(methylamino)ethyl]-3,7-diazabicyclo[3.3.1]non-3-yl}-1-oxopropan-2-yl]-2-oxopy rrolidin-3-yl}naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Factor Xa complex with GTC000441
to be published
|
|
5I3U
 
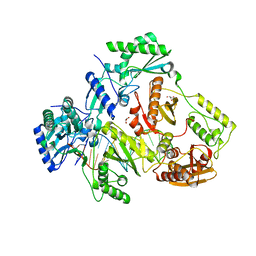 | |
1CDP
 
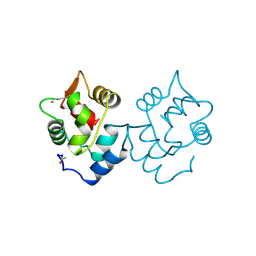 | |
3SSF
 
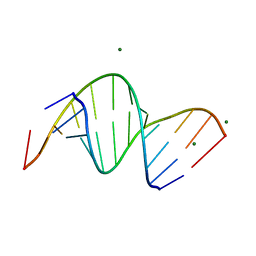 | | Crystal structure of RNA:DNA dodecamer corresponding to HIV-1 polypurine tract, at 1.6 A resolution. | | Descriptor: | 5'-D(*CP*CP*TP*TP*TP*TP*CP*TP*TP*TP*TP*A)-3', 5'-R(*UP*AP*AP*AP*AP*GP*AP*AP*AP*AP*GP*G)-3', MAGNESIUM ION | | Authors: | Drozdzal, P, Michalska, K, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2011-07-08 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA/DNA dodecamer corresponding to the HIV-1 polypurine tract at 1.6 Angstrom resolution
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4J6X
 
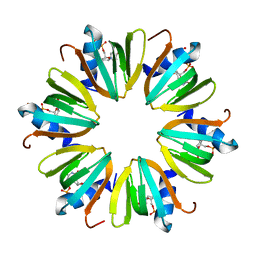 | |
1ANV
 
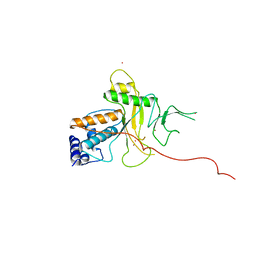 | | ADENOVIRUS 5 DBP/URANYL FLUORIDE SOAK | | Descriptor: | ADENOVIRUS SINGLE-STRANDED DNA-BINDING PROTEIN, URANYL (VI) ION, ZINC ION | | Authors: | Kanellopoulos, P.N, Tsernoglou, D, Van Der Vliet, P.C, Tucker, P.A. | | Deposit date: | 1996-03-14 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational change of the adenovirus DNA-binding protein induced by soaking crystals with K3UO2F5 solutions.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1BBU
 
 | | LYSYL-TRNA SYNTHETASE (LYSS) COMPLEXED WITH LYSINE | | Descriptor: | LYSINE, PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
4J6Y
 
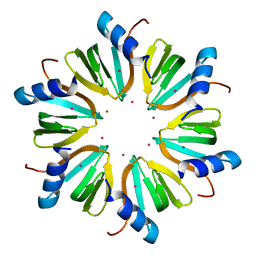 | |
1QB1
 
 | |
1QQL
 
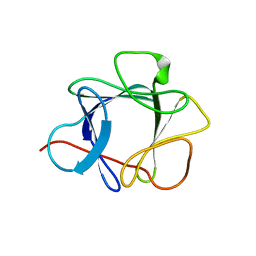 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7/1 CHIMERA | | Descriptor: | FIBROBLAST GROWTH FACTOR 7/1 CHIMERA | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
3THK
 
 | | Structure of SH3 chimera with a type II ligand linked to the chain C-terminal | | Descriptor: | BETA-MERCAPTOETHANOL, Proline-rich peptide, SULFATE ION, ... | | Authors: | Gabdulkhakov, A.G, Gushchina, L.V, Nikulin, A.D, Nikonov, S.V, Filimonov, V.V. | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structure of Spectrin SH3 Domain Fused with a Proline-Rich Peptide.
J.Biomol.Struct.Dyn., 29, 2011
|
|
4J6W
 
 | | Crystal structure of HFQ from Pseudomonas aeruginosa in complex with CTP | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Nikulin, A.D, Murina, V, Lekontseva, N. | | Deposit date: | 2013-02-12 | | Release date: | 2013-07-31 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1QQK
 
 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7 (KERATINOCYTE GROWTH FACTOR) | | Descriptor: | FIBROBLAST GROWTH FACTOR 7 | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
1ER8
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | Endothiapepsin, H-77 | | Authors: | Hemmings, A.M, Veerapandian, B, Szelke, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1989-10-16 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
2IXD
 
 | | Crystal structure of the putative deacetylase BC1534 from Bacillus cereus | | Descriptor: | ACETATE ION, LMBE-RELATED PROTEIN, ZINC ION | | Authors: | Fadouloglou, V.E, Bouriotis, V, Kokkinidis, M. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Bczbp, a Zinc-Binding Protein from Bacillus Cereus
FEBS J., 274, 2007
|
|
1HIO
 
 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN, ALPHA CARBONS ONLY | | Descriptor: | HISTONE H2A, HISTONE H2B, HISTONE H3, ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1991-09-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
2IU0
 
 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 1,5-DIHYDROIMIDAZO[4,5-C][1,2,6]THIADIAZIN-4(3H)-ONE 2,2-DIOXIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, POTASSIUM ION | | Authors: | Xu, L, Chong, Y, Hwang, I, Onofrio, A.D, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2006-05-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-Based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase
J.Biol.Chem., 282, 2007
|
|
