8SUZ
 
 | | Open State of the SARS-CoV-2 Envelope Protein Transmembrane Domain, Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Medeiros-Silva, J, Dregni, A.J, Somberg, N.H, Hong, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure of the open SARS-CoV-2 E viroporin.
Sci Adv, 9, 2023
|
|
4ISR
 
 | | Binding domain of Botulinum neurotoxin DC in complex with rat synaptotagmin II | | Descriptor: | Neurotoxin, SULFATE ION, Synaptotagmin-2 | | Authors: | Berntsson, R.P.-A, Peng, L, Svensson, L.M, Dong, M, Stenmark, P. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin DC in Complex with Its Protein Receptors Synaptotagmin I and II.
Structure, 21, 2013
|
|
4ISQ
 
 | | Binding domain of Botulinum neurotoxin DC in complex with human synaptotagmin I | | Descriptor: | GLYCEROL, Neurotoxin, SULFATE ION, ... | | Authors: | Berntsson, R.P.-A, Peng, L, Svensson, L.M, Dong, M, Stenmark, P. | | Deposit date: | 2013-01-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Botulinum Neurotoxin DC in Complex with Its Protein Receptors Synaptotagmin I and II.
Structure, 21, 2013
|
|
8TYX
 
 | |
8TTN
 
 | | PHF1-Phosphomimetic Tau Filaments (Full-length, Cofactor-Free 0N4R Tau S396E, S400E, T403E, S404E) | | Descriptor: | Microtubule-associated protein tau | | Authors: | El Mammeri, N, Dregni, A.J, Duan, P, Hong, M. | | Deposit date: | 2023-08-14 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structures of AT8 and PHF1 phosphomimetic tau: Insights into the posttranslational modification code of tau aggregation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TTL
 
 | | AT8-Phosphomimetic Tau Filaments (Full-length, Cofactor-Free 0N4R Tau S202E, T205E, S208E) | | Descriptor: | Microtubule-associated protein tau | | Authors: | El Mammeri, N, Dregni, A.J, Duan, P, Hong, M. | | Deposit date: | 2023-08-14 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of AT8 and PHF1 phosphomimetic tau: Insights into the posttranslational modification code of tau aggregation.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8TYY
 
 | |
4KBB
 
 | | Structure of Botulinum neurotoxin B binding domain in complex with both synaptotagmin II and GD1a | | Descriptor: | Botulinum neurotoxin type B, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Berntsson, R.P.A, Peng, L, Dong, M, Stenmark, P. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of dual receptor binding to botulinum neurotoxin B.
Nat Commun, 4, 2013
|
|
7JK8
 
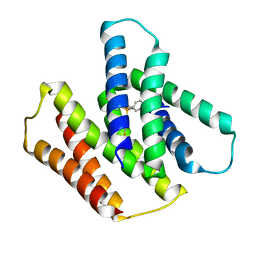 | | EmrE S64V mutant bound to tetra(4-fluorophenyl)phosphonium at pH 5.8 | | Descriptor: | Multidrug SMR transporter, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Hisao, G, Mandala, V.S, Thomas, N.E, Soltani, M, Salter, E.A, Davis Jr, J.H, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers.
Nat Commun, 12, 2021
|
|
5JMC
 
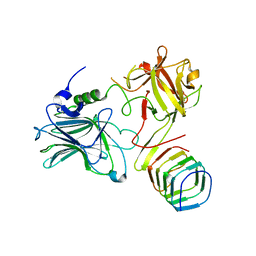 | | Receptor binding domain of Botulinum neurotoxin A in complex with rat SV2C | | Descriptor: | Botulinum neurotoxin type A, Synaptic vesicle glycoprotein 2C | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2KQT
 
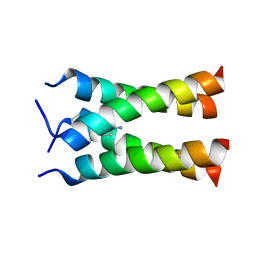 | | Solid-state NMR structure of the M2 transmembrane peptide of the influenza A virus in DMPC lipid bilayers bound to deuterated amantadine | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, M2 protein | | Authors: | Cady, S.D, Schmidt-Rohr, K, Wang, J, Soto, C.S, DeGrado, W.F, Hong, M. | | Deposit date: | 2009-11-18 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-08 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers
Nature, 463, 2010
|
|
2M35
 
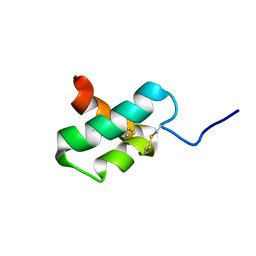 | | NMR study of k-Ssm1a | | Descriptor: | k-Ssm1a | | Authors: | King, G.F, Undheim, E.A, Mobli, M, Yang, S, Rong, M, Lai, R. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR study of k-Ssm1a
To be Published
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
5WYQ
 
 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
5WYR
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
2XJG
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 1,3-DIHYDROISOINDOL-2-YL-(2-HYDROXY-4-METHOXY-5-PROPAN-2-YL-PHENYL)METHANONE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, B, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XJJ
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 1,3-DIHYDROISOINDOL-2-YL-(6-HYDROXY-3,3-DIMETHYL-1,2-DIHYDROINDOL-5-YL)METHANONE, GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XJX
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, Onalespib | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XAB
 
 | | Structure of HSP90 with an inhibitor bound | | Descriptor: | 4-(1,3-DIHYDRO-2H-ISOINDOL-2-YLCARBONYL)-6-(1-METHYLETHYL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, m, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, O'Brian, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2MUZ
 
 | | ssNMR structure of a designed rocker protein | | Descriptor: | designed rocker protein | | Authors: | Wang, T, Joh, N, Wu, Y, DeGrado, W.F, Hong, M. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2015-01-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a transmembrane Zn2+-transporting four-helix bundle.
Science, 346, 2014
|
|
4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
2XHR
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 4-CHLORO-6-[2,4-DICHLORO-5-(2-MORPHOLIN-4-YLETHOXY)PHENYL]PYRIMIDIN-2-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
2XHX
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 2-TERT-BUTYL-4-(1,3-DIHYDRO-2H-ISOINDOL-2-YLCARBONYL)PHENOL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-06-21 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
2XDS
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 1-CHLORO-4-METHYLPHTHALAZINE, DIMETHYL SULFOXIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
6JKI
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 25, 2019
|
|
