1TYZ
 
 | | Crystal structure of 1-Aminocyclopropane-1-carboyxlate Deaminase from Pseudomonas | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-08 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes:Insight into the mechanism of unique pyrodoxial-5'-phosphate dependent cyclopropane ring opening reaction
Biochemistry, 43, 2004
|
|
1TZK
 
 | | Crystal structure of 1-aminocyclopropane-1-carboxylate-deaminase complexed with alpha-keto-butyrate | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 2-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
2VJO
 
 | | Formyl-CoA transferase mutant variant Q17A with aspartyl-CoA thioester intermediates and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, COENZYME A, ... | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2VDG
 
 | | Barley Aldose Reductase 1 complex with butanol | | Descriptor: | 1-BUTANOL, ALDOSE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2007-10-08 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2VBF
 
 | | The holostructure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis | | Descriptor: | BRANCHED-CHAIN ALPHA-KETOACID DECARBOXYLASE, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Berthold, C.L, Gocke, D, Wood, M.D, Leeper, F, Pohl, M, Schneider, G. | | Deposit date: | 2007-09-12 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Branched-Chain Keto Acid Decarboxylase (Kdca) from Lactococcus Lactis Provides Insights Into the Structural Basis for the Chemo- and Enantioselective Carboligation Reaction
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2UWP
 
 | | Factor Xa inhibitor complex | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHANESULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Young, R.J, Brown, D, Burns-Kurtis, C.L, Chan, C, Convery, M.A, Hubbard, J.A, Kelly, H.A, Pateman, A.J, Patikis, A, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Zhou, P, Thorpe, J.H. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Selective and Dual Action Orally Active Inhibitors of Thrombin and Factor Xa.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2VCS
 
 | | Structure of isoniazid (INH) bound to cytosolic soybean ascorbate peroxidase mutant H42A | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-09-26 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
2V3W
 
 | | Crystal structure of the benzoylformate decarboxylase variant L461A from Pseudomonas putida | | Descriptor: | BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Gocke, D, Walter, L, Gauchenova, K, Kolter, G, Knoll, M, Berthold, C.L, Schneider, G, Pleiss, J, Mueller, M, Pohl, M. | | Deposit date: | 2007-06-25 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Protein Design of Thdp-Dependent Enzymes-Engineering Stereoselectivity.
Chembiochem, 9, 2008
|
|
2V23
 
 | | Structure of cytochrome c peroxidase mutant N184R Y36A | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-05-31 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
2VJL
 
 | | Formyl-CoA transferase with aspartyl-CoA thioester intermediate derived from formyl-CoA | | Descriptor: | CHLORIDE ION, COENZYME A, FORMYL-COENZYME A TRANSFERASE, ... | | Authors: | Berthold, C.L, Toyota, C.G, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2007-12-11 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reinvestigation of the Catalytic Mechanism of Formyl-Coa Transferase, a Class III Coa-Transferase.
J.Biol.Chem., 283, 2008
|
|
2WOC
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1TRO
 
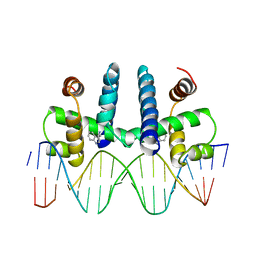 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
1TZM
 
 | | Crystal structure of ACC deaminase complexed with substrate analog b-chloro-D-alanine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 3-chloro-D-alanine, AMINO-ACRYLATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
2VOF
 
 | | Structure of mouse A1 bound to the Puma BH3-domain | | Descriptor: | BCL-2-BINDING COMPONENT 3, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2W2G
 
 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
1TZ2
 
 | | Crystal structure of 1-aminocyclopropane-1-carboyxlate deaminase complexed with ACC | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1U34
 
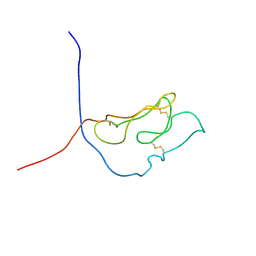 | | 3D NMR structure of the first extracellular domain of CRFR-2beta, a type B1 G-protein coupled receptor | | Descriptor: | Corticotropin releasing factor receptor 2 | | Authors: | Grace, C.R, Perrin, M.H, DiGruccio, M.R, Miller, C.L, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR structure and peptide hormone binding site of the first extracellular domain of a type B1 G protein-coupled receptor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TZJ
 
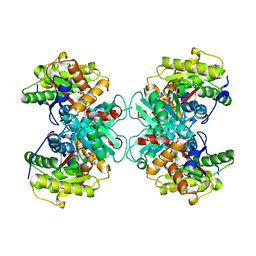 | | Crystal Structure of 1-aminocyclopropane-1-carboxylate deaminase complexed with d-vinyl glycine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, D-VINYLGLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
1U65
 
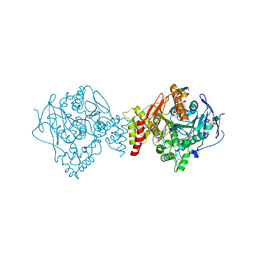 | | Ache W. CPT-11 | | Descriptor: | (4S)-4,11-DIETHYL-4-HYDROXY-3,14-DIOXO-3,4,12,14-TETRAHYDRO-1H-PYRANO[3',4':6,7]INDOLIZINO[1,2-B]QUINOLIN-9-YL 1,4'-BIPIPERIDINE-1'-CARBOXYLATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Harel, M, Hyatt, J.L, Brumshtein, B, Morton, C.L, Wadkins, R.W, Silman, I, Sussman, J.L, Potter, P.M, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2004-07-29 | | Release date: | 2005-07-19 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The crystal structure of the complex of the anticancer prodrug 7-ethyl-10-[4-(1-piperidino)-1-piperidino]-carbonyloxycamptothecin (CPT-11) with Torpedo californica acetylcholinesterase provides a molecular explanation for its cholinergic action
Mol.Pharmacol., 67, 2005
|
|
2VOG
 
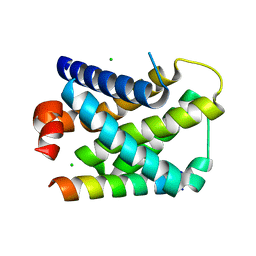 | | Structure of mouse A1 bound to the Bmf BH3-domain | | Descriptor: | BCL-2-MODIFYING FACTOR, BCL-2-RELATED PROTEIN A1, CHLORIDE ION | | Authors: | Smits, C, Czabotar, P.E, Hinds, M.G, Day, C.L. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity Underpins Promiscuous Binding of the Prosurvival Protein A1.
Structure, 16, 2008
|
|
2WBM
 
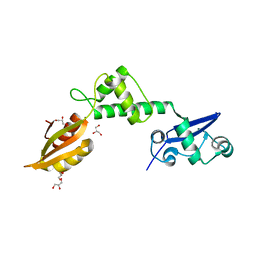 | | Crystal structure of mthSBDS, the homologue of the Shwachman-Bodian- Diamond syndrome protein in the euriarchaeon Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, GLYCEROL, RIBOSOME MATURATION PROTEIN SDO1 HOMOLOG, ... | | Authors: | Ng, C.L, Isupov, M.N, Lebedev, A.A, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-03-02 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Flexibility and Molecular Interactions of an Archaeal Homologue of the Shwachman-Bodian-Diamond Syndrome Protein.
Bmc Struct.Biol., 9, 2009
|
|
2XUC
 
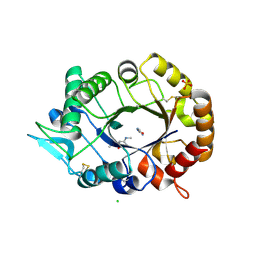 | | Natural product-guided discovery of a fungal chitinase inhibitor | | Descriptor: | 1-methyl-3-(N-methylcarbamimidoyl)urea, CHITINASE, CHLORIDE ION, ... | | Authors: | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2010-10-18 | | Release date: | 2010-10-27 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
2XHY
 
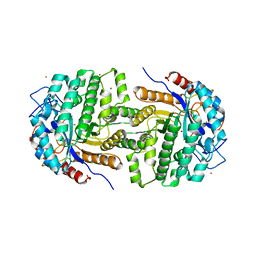 | | Crystal Structure of E.coli BglA | | Descriptor: | 6-PHOSPHO-BETA-GLUCOSIDASE BGLA, BROMIDE ION, SULFATE ION | | Authors: | Totir, M, Zubieta, C, Echols, N, May, A.P, Gee, C.L, nanao, M, alber, T. | | Deposit date: | 2010-06-24 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
2WOE
 
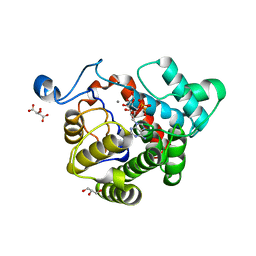 | | Crystal Structure of the D97N variant of dinitrogenase reductase- activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP-ribose | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOD
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP- ribsoyllysine | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
