5ZH0
 
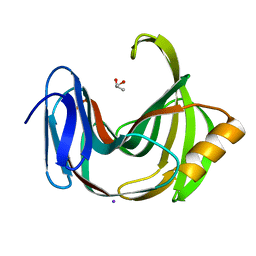 | | Crystal Structures of Endo-beta-1,4-xylanase II | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Zhang, X, Wan, Q, Li, Z. | | Deposit date: | 2018-03-10 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal Structures of Endo-beta-1,4-xylanase II
To be published
|
|
5ZF3
 
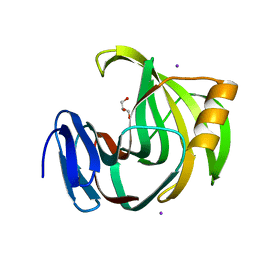 | | Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Zhang, X, Wan, Q, Li, Z. | | Deposit date: | 2018-03-02 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose
To be published
|
|
5Z93
 
 | |
5XJC
 
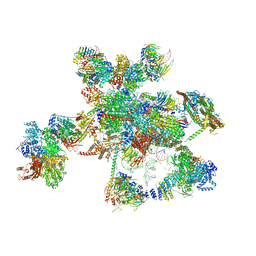 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
6LSV
 
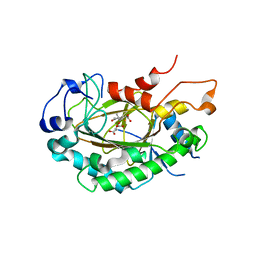 | | Crystal structure of JOX2 in complex with 2OG, Fe, and JA | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Probable 2-oxoglutarate-dependent dioxygenase At5g05600, ... | | Authors: | Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure-guided analysis of Arabidopsis JASMONATE-INDUCED OXYGENASE (JOX) 2 reveals key residues for recognition of jasmonic acid substrate by plant JOXs.
Mol Plant, 14, 2021
|
|
5Z94
 
 | |
5ZE3
 
 | |
5ZH9
 
 | |
8IEW
 
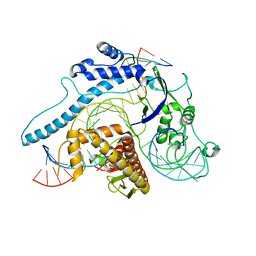 | | Cas005-crRNA-DNA complex | | Descriptor: | Cas005, DNA (55-MER), RNA (38-MER) | | Authors: | Zhang, X, Duan, Z.Q, Zhu, J.K. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of Cas005-crRNA-DNA complex at 3.1 Angstroms resolution
To Be Published
|
|
7YC8
 
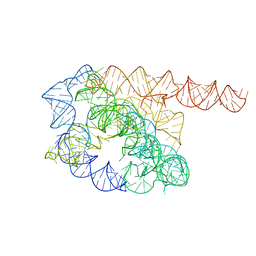 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (388-MER) | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCI
 
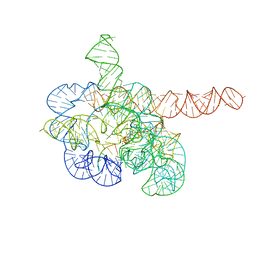 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (389-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCG
 
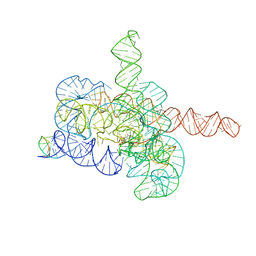 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
7YCH
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8JT4
 
 | |
8JSD
 
 | | Alginate lyase mutant-D180G | | Descriptor: | Alginate lyase | | Authors: | Zhang, X, Pan, L.X, Yang, D.F. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.414 Å) | | Cite: | Substrate-binding mode guided protein design of alginate lyase FlAlyA for altered end-product distribution
To Be Published
|
|
8HYR
 
 | |
7XGD
 
 | | Cryo-EM structure of Apo-IGF1R map 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin-like growth factor 1 receptor | | Authors: | Zhang, X, Wu, C. | | Deposit date: | 2022-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of Apo-IGF1R
To Be Published
|
|
7DVG
 
 | |
3VNB
 
 | |
3VNA
 
 | |
6X1A
 
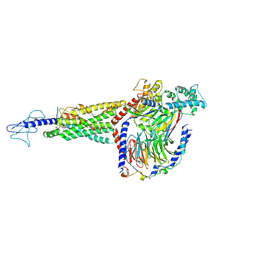 | | Non peptide agonist PF-06882961, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 2-[(4-{6-[(4-cyano-2-fluorophenyl)methoxy]pyridin-2-yl}piperidin-1-yl)methyl]-1-{[(2S)-oxetan-2-yl]methyl}-1H-benzimidazole-6-carboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X19
 
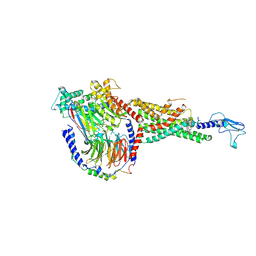 | | Non peptide agonist CHU-128, bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | 3-{(1S)-1-[5-(2,2-dimethylmorpholin-4-yl)-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl]butyl}-1,2,4-oxadiazol-5(4H)-one, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
6X18
 
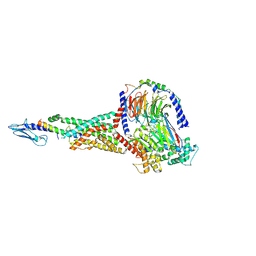 | | GLP-1 peptide hormone bound to Glucagon-Like peptide-1 (GLP-1) Receptor | | Descriptor: | Glucagon, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Zhang, X, Danev, R. | | Deposit date: | 2020-05-18 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Differential GLP-1R Binding and Activation by Peptide and Non-peptide Agonists.
Mol.Cell, 80, 2020
|
|
1J04
 
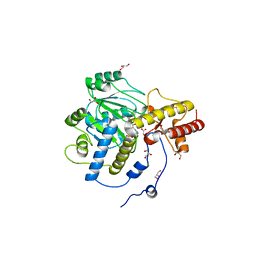 | | Structural mechanism of enzyme mistargeting in hereditary kidney stone disease in vitro | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, alanine--glyoxylate aminotransferase | | Authors: | Zhang, X, Djordjevic, S, Bartlam, M, Ye, S, Rao, Z, Danpure, C.J. | | Deposit date: | 2002-10-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural implications of a G170R mutation of alanine:glyoxylate aminotransferase that is associated with peroxisome-to-mitochondrion mistargeting.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7WOP
 
 | | The local refined map of Omicron spike with bispecific antibody FD01 | | Descriptor: | 16L9, GW01, Spike protein S1 | | Authors: | Zhang, X, Zhan, W.Q, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Combating the SARS-CoV-2 Omicron (BA.1) and BA.2 with potent bispecific antibodies engineered from non-Omicron neutralizing antibodies
Cell Discov, 8, 2022
|
|
