6KI1
 
 | |
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
8DWB
 
 | |
6KI2
 
 | |
6KIK
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor tolrestat | | Descriptor: | Oxidoreductase, aldo/keto reductase family, TOLRESTAT | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KIY
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | Authors: | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | Deposit date: | 2019-07-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
6KY6
 
 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | Descriptor: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | Descriptor: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | Descriptor: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | Descriptor: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
5D7C
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[1-(pyridin-2-yl)-6-(pyridin-3-yl)-1H-pyrrolo[3,2-b]pyridin-3-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
3C0F
 
 | | Crystal Structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source | | Descriptor: | Uncharacterized protein AF_1514 | | Authors: | Li, Y, Bahti, P, Shaw, N, Song, G, Yin, J, Zhu, J.-Y, Zhang, H, Xu, H, Wang, B.-C, Liu, Z.-J. | | Deposit date: | 2008-01-20 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel non-Pfam protein AF1514 from Archeoglobus fulgidus DSM 4304 solved by S-SAD using a Cr X-ray source.
Proteins, 71, 2008
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hong, S, Horton, J.R, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
7MHU
 
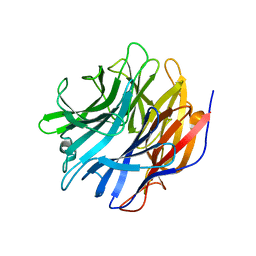 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
1YFL
 
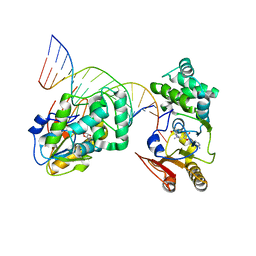 | | T4Dam in Complex with Sinefungin and 16-mer Oligonucleotide Showing Semi-specific and Specific Contact and Flipped Base | | Descriptor: | 5'-D(P*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*GP*A)-3', DNA adenine methylase, SINEFUNGIN | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
5T0U
 
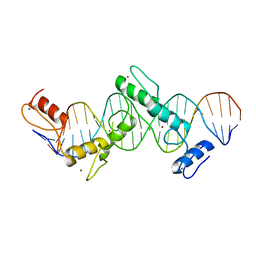 | | CTCF ZnF2-7 and DNA complex structure | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.199 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
6M7I
 
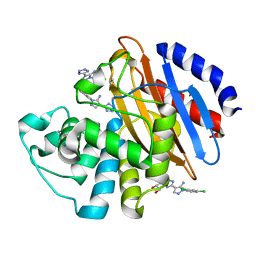 | | Crystal structure of KPC-2 with compound 3 | | Descriptor: | 1-(2,4-dichlorophenyl)-4-(1H-tetrazol-5-yl)-1H-pyrazol-5-amine, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
5T00
 
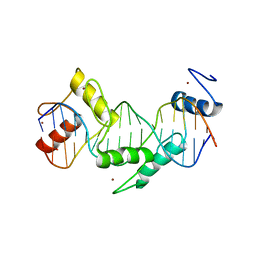 | | Human CTCF ZnF3-7 and methylated DNA complex | | Descriptor: | DNA (5'-GCCAGCAGGGGG(5CM)GCTA-3'), DNA (5'-TAG(5CM)GCCCCCTGCTGGC-3'), Transcriptional repressor CTCF, ... | | Authors: | Hashimoto, H, Wang, D, Cheng, X. | | Deposit date: | 2016-08-15 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
8W4V
 
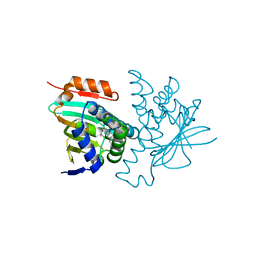 | | Crystal structure of human HSP90 in complex with compound 4 | | Descriptor: | 4-[2-[(dimethylamino)methyl]phenyl]sulfanylbenzene-1,3-diol, Heat shock protein HSP 90-alpha, MAGNESIUM ION | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-08-25 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
7L5M
 
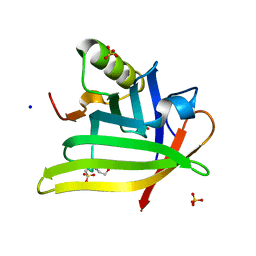 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
7L5L
 
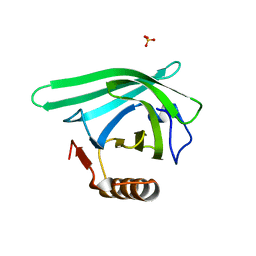 | |
