5YNQ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YOU
 
 | | Crystal structure of BRD4-BD1 bound with hjp64 | | Descriptor: | (3~{R})-4-cyclopropyl-~{N},1,3-trimethyl-~{N}-(4-methylphenyl)-2-oxidanylidene-3~{H}-quinoxaline-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Bing, X, Jianping, H, Yanlian, L, Danyan, C. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Crystal structure of BRD4-BD1 bound with hjp64
To Be Published
|
|
5YN5
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex | | Descriptor: | ZINC ION, nsp10 protein, nsp16 protein | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNF
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNP
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to sinefungin and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNB
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to Sinefungin | | Descriptor: | SINEFUNGIN, ZINC ION, nsp10 protein, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNM
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
4JZR
 
 | | Structure of Prolyl Hydroxylase Domain-containing Protein (PHD) with Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-(biphenyl-4-yl)-8-[(1-methyl-1H-imidazol-2-yl)methyl]-2,8-diazaspiro[4.5]decan-1-one, Egl nine homolog 1, ... | | Authors: | Ma, Y, Yang, L. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel complex crystal structure of prolyl hydroxylase domain-containing protein 2 (PHD2): 2,8-Diazaspiro[4.5]decan-1-ones as potent, orally bioavailable PHD2 inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
5YAW
 
 | |
2XKX
 
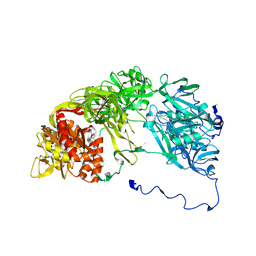 | | Single particle analysis of PSD-95 in negative stain | | Descriptor: | DISKS LARGE HOMOLOG 4 | | Authors: | Fomina, S, Howard, T.D, Sleator, O.K, Golovanova, M, O'Ryan, L, Leyland, M.L, Grossmann, J.G, Collins, R.F, Prince, S.M. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (22.9 Å), SOLUTION SCATTERING | | Cite: | Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Biochim.Biophys.Acta, 1808, 2011
|
|
6KF1
 
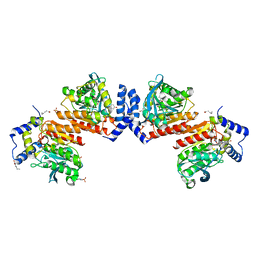 | | Microbial Hormone-sensitive lipase- E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEXANOIC ACID, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Functional and Structural Insights into Environmental Adaptation of a Novel Hormone-sensitive Lipase, E53, Obtained from Erythrobacter longus
To Be Published
|
|
5YNO
 
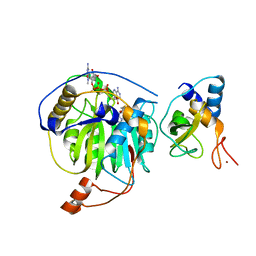 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
6J3L
 
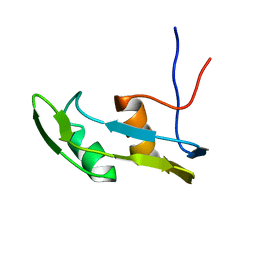 | | Solution structure of the N-terminal extended protuberant domain of eukaryotic ribosomal stalk protein P0 | | Descriptor: | 60S acidic ribosomal protein P0 | | Authors: | Choi, K.H.A, Lee, K.M, Yang, L, Wing-Heng Yu, C, Banfield, D.K, Ito, K, Uchiumi, T, Wong, K.B. | | Deposit date: | 2019-01-04 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mutagenesis Studies Evince the Role of the Extended Protuberant Domain of Ribosomal Protein uL10 in Protein Translation.
Biochemistry, 58, 2019
|
|
2XKY
 
 | | Single particle analysis of Kir2.1NC_4 in negative stain | | Descriptor: | INWARD RECTIFIER POTASSIUM CHANNEL 2 | | Authors: | Fomina, S, Howard, T.D, Sleator, O.K, Golovanova, M, O'Ryan, L, Leyland, M.L, Grossmann, J.G, Collins, R.F, Prince, S.M. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17.200001 Å), SOLUTION SCATTERING | | Cite: | Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Biochim.Biophys.Acta, 1808, 2011
|
|
1XI2
 
 | | Quinone Reductase 2 in Complex with Cancer Prodrug CB1954 | | Descriptor: | 5-(AZIRIDIN-1-YL)-2,4-DINITROBENZAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, NRH dehydrogenase [quinone] 2, ... | | Authors: | Fu, Y, Buryanovskyy, L, Zhang, Z. | | Deposit date: | 2004-09-21 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of quinone reductase 2 in complex with cancer prodrug CB1954
Biochem.Biophys.Res.Commun., 336, 2005
|
|
4Q7H
 
 | |
7DYQ
 
 | | Crystal structure of histone lysine demethylase 4D (KDM4D) in complex with the inhibitor 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile | | Descriptor: | 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile, FE (III) ION, Lysine-specific demethylase 4D | | Authors: | Wang, T, Yang, L. | | Deposit date: | 2021-01-22 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of histone lysine demethylase 4D (KDM4D) in complex with the inhibitor 5-hydroxy-2-methylpyrazolo[1,5-a]pyrido[3,2-e]pyrimidine-3-carbonitrile
To Be Published
|
|
7DYG
 
 | |
7EGQ
 
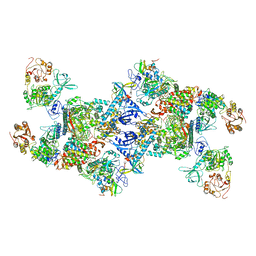 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
3UIM
 
 | |
8GWO
 
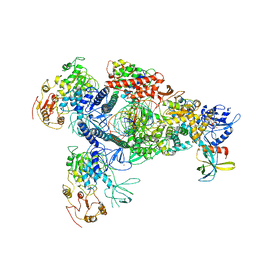 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
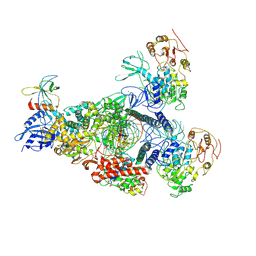 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
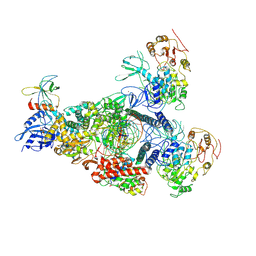 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWB
 
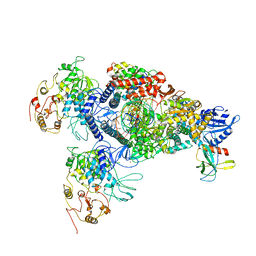 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
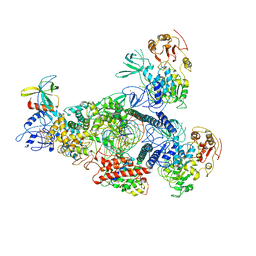 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
