8IHN
 
 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHT
 
 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
6OQX
 
 | | Human Liver Receptor Homolog-1 bound to the agonist 5N and a fragment of the Tif2 coregulator | | Descriptor: | (8beta,11alpha,12alpha)-8-(1-phenylethenyl)-1,6:7,14-dicycloprosta-1,3,5,7(14)-tetraen-11-yl sulfamate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
6OR1
 
 | | Human LRH-1 bound to the agonist 2N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]acetamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
6OQY
 
 | | Human LRH-1 bound to the agonist 6N and a fragment of the Tif2 coregulator | | Descriptor: | N-[(1S,3aR,6aR)-5-hexyl-4-phenyl-3a-(1-phenylethenyl)-1,2,3,3a,6,6a-hexahydropentalen-1-yl]sulfuric diamide, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | Mays, S.G, Ortlund, E.A. | | Deposit date: | 2019-04-29 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Development of the First Low Nanomolar Liver Receptor Homolog-1 Agonist through Structure-guided Design.
J.Med.Chem., 62, 2019
|
|
4RVG
 
 | | Crystal structure of MtmC in complex with SAM and TDP | | Descriptor: | ACETATE ION, D-mycarose 3-C-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVD
 
 | | Crystal structure of MtmC in complex with SAM | | Descriptor: | ACETATE ION, D-mycarose 3-C-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RVF
 
 | | Crystal structure of MtmC in complex with TDP | | Descriptor: | D-mycarose 3-C-methyltransferase, THYMIDINE-5'-DIPHOSPHATE, ZINC ION | | Authors: | Tsodikov, O.V, Hou, C, Chen, J.-M, Rohr, J. | | Deposit date: | 2014-11-26 | | Release date: | 2015-01-28 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
4RV9
 
 | | Crystal structure of MtmC in complex with SAH | | Descriptor: | ACETATE ION, CHLORIDE ION, D-mycarose 3-C-methyltransferase, ... | | Authors: | Hou, C, Chen, J.-M, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2014-11-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into MtmC, a Bifunctional Ketoreductase-Methyltransferase Involved in the Assembly of the Mithramycin Trisaccharide Chain.
Biochemistry, 54, 2015
|
|
6H02
 
 | |
6PLI
 
 | | Crystal Structure of EcDsbA in a complex with purified oxadiazole 11 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-methyl-N-[2-(5-methyl-1,2,4-oxadiazol-3-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-07-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PC9
 
 | | Crystal Structure of EcDsbA in a complex with purified methylpiperazinone 6 | | Descriptor: | 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiLX).
J.Med.Chem., 63, 2020
|
|
6PIQ
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product G6 (pyrazole 9) | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG2
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product H5 (morpholine 8) | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PBI
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine 8 | | Descriptor: | 2-methyl-4-{4-[2-(morpholin-4-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PGJ
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product A5 (Morpholine carboxylic acid 7) | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, (3S)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PDH
 
 | | Crystal Structure of EcDsbA in a complex with purified pyrazole 9 | | Descriptor: | 2-[4-(4-cyano-3-methylphenoxy)phenyl]-N-ethyl-N-[2-(1H-pyrazol-1-yl)ethyl]acetamide, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-19 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PG1
 
 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product F1 (methylpiperazinone 6) | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-4-{4-[2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl]phenoxy}benzonitrile, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6PD7
 
 | | Crystal Structure of EcDsbA in a complex with purified morpholine carboxylic acid 7 | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-18 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
6XD1
 
 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-640 | | Descriptor: | (2R)-4-(butyl{[2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}carbamoyl)-1-(2,2-diphenylpropanoyl)piperazine-2-carboxylic acid, RNA-dependent RNA polymerase, ZINC ION | | Authors: | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
6XD0
 
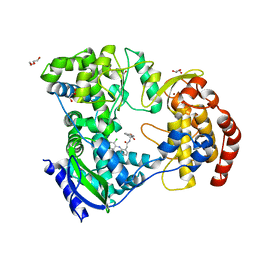 | | Dengue serotype 3 RNA-dependent RNA polymerase bound to NITD-434 | | Descriptor: | 2-[({2-[(2,6-dichlorophenyl)amino]phenyl}acetyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Arora, R, Benson, T.E, Liew, C.W, Lescar, J. | | Deposit date: | 2020-06-09 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies.
J.Virol., 94, 2020
|
|
5YM8
 
 | |
5YM6
 
 | |
