2QPK
 
 | | Crystal structure of the complex of bovine lactoperoxidase with salicylhydroxamic acid at 2.34 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-07-24 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Biochim.Biophys.Acta, 1865, 2017
|
|
3JQ5
 
 | | Phospholipase A2 Prevents the Aggregation of Amyloid Beta Peptides: Crystal Structure of the Complex of Phospholipase A2 with Octapeptide Fragment of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser at 2 A Resolution | | Descriptor: | Amyloid Beta Peptide, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-09-06 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phospholipase A2 Prevents the Aggregation of Amyloid Beta Peptides: Crystal Structure of the Complex of Phospholipase A2 with Octapeptide Fragment of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser at 2 A Resolution
To be Published
|
|
3NW3
 
 | | Crystal structure of the complex of peptidoglycan recognition protein (PGRP-S) with the PGN Fragment at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ALANINE, D-GLUTAMINE, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiligand specificity of pathogen-associated molecular pattern-binding site in peptidoglycan recognition protein
J.Biol.Chem., 286, 2011
|
|
1YXL
 
 | | Crystal structure of a novel phospholipase A2 from Naja naja sagittifera at 1.5 A resolution | | Descriptor: | ACETIC ACID, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Singh, R.K, Jabeen, T, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-02-22 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Crystal Structure of a novel phospholipase A2 from Naja naja sagittifera at 1.5 A resolution
To be Published
|
|
5H16
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with citrate at 2.3 A resolution. | | Descriptor: | CITRIC ACID, Phosphopantetheine adenylyltransferase | | Authors: | Gupta, A, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-10-08 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii at 2.3 A resolution.
To Be Published
|
|
5YLA
 
 | |
5YL8
 
 | | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, SODIUM ION | | Authors: | Bairagya, H.R, Sharma, P, Iqbal, N, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-10-17 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of inactive dimeric peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.79 A resolution
To Be Published
|
|
1ZR8
 
 | | Crystal Structure of the complex formed between group II phospholipase A2 and a plant alkaloid ajmaline at 2.0A resolution | | Descriptor: | ACETIC ACID, AJMALINE, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Mahendra, M, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2005-05-19 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of the complex formed between group II phospholipase A2 and a plant alkaloid ajmaline at 2.0A resolution
To be Published
|
|
5YN4
 
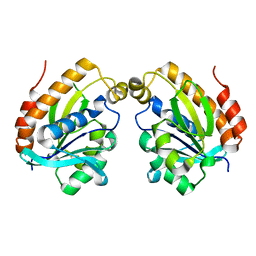 | |
2B65
 
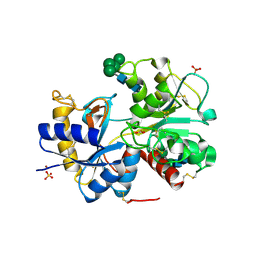 | | Crystal structure of the complex of C-lobe of bovine lactoferrin with maltose at 1.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Singh, N, Prem kumar, R, Jabeen, T, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2005-09-30 | | Release date: | 2005-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the complex of C-lobe of bovine lactoferrin with maltose at 1.5A resolution
To be published
|
|
1ZWP
 
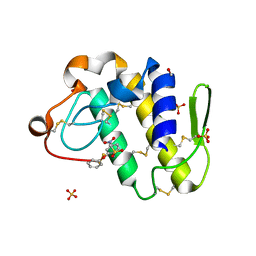 | | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2 | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2
To be Published
|
|
5ILX
 
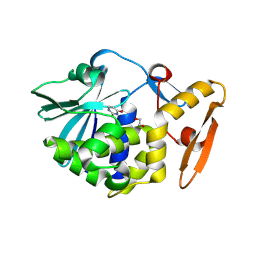 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
5ILW
 
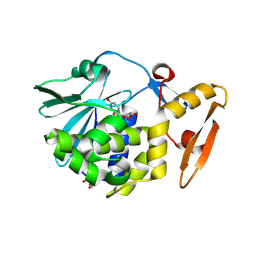 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with Uridine at 1.97 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
1ZBC
 
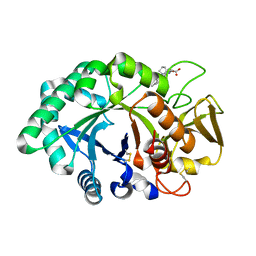 | | Crystal Structure of the porcine signalling protein liganded with the peptide Trp-Pro-Trp (WPW) at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3 mer peptide, signal processing protein | | Authors: | Srivastava, D.B, Kaur, P, Kumar, J, Somvanshi, R.K, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of the porcine signalling protein liganded with the peptide Trp-Pro-Trp (WPW) at 2.3 A resolution
To be Published
|
|
1ZB5
 
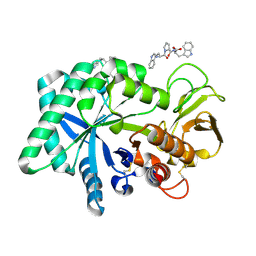 | | Recognition of peptide ligands by signalling protein from porcine mammary gland (SPP-40): Crystal structure of the complex of SPP-40 with a peptide Trp-Pro-Trp at 2.45A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PEPTIDE TRP-PRO-TRP, signal processing protein | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Somvanshi, R.K, Sharma, S, Dey, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-04-07 | | Release date: | 2005-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Recognition of peptide ligands by signalling protein from porcine mammary gland (SPP-40): Crystal structure of the complex of SPP-40 with a peptide Trp-Pro-Trp at 2.45A resolution
To be Published
|
|
5HBC
 
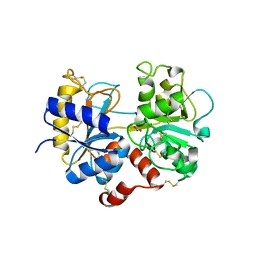 | | Intermediate structure of iron-saturated C-lobe of bovine lactoferrin at 2.79 Angstrom resolution indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-31 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron saturated C-lobe of bovine lactoferrin at pH 6.8 indicates a weakening of iron coordination
Proteins, 84, 2016
|
|
5ZXM
 
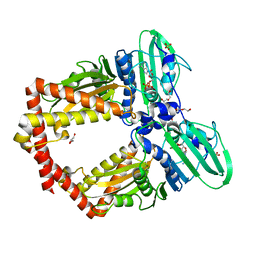 | | Crystal Structure of GyraseB N-terminal at 1.93A Resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA gyrase subunit B, ... | | Authors: | Tiwari, P, Gupta, D, Sachdeva, E, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural insights into the transient closed conformation and pH dependent ATPase activity of S.Typhi GyraseB N- terminal domain.
Arch.Biochem.Biophys., 701, 2021
|
|
4OPP
 
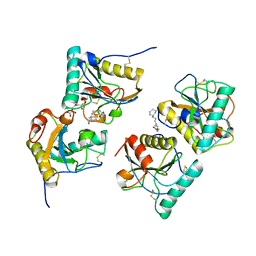 | | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution | | Descriptor: | 11-cyclohexylundecanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-06 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution
To be Published
|
|
4ORV
 
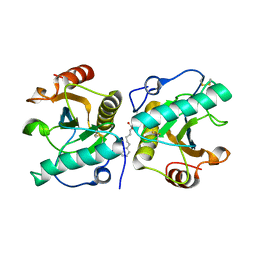 | | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-phenylheptanoic acid, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution
To be Published
|
|
5Z4V
 
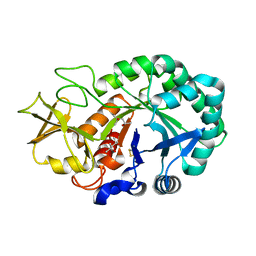 | | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Sharma, P, Singh, P.K, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2018-01-15 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40
To Be Published
|
|
2AYW
 
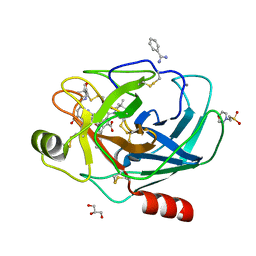 | | Crystal Structure of the complex formed between trypsin and a designed synthetic highly potent inhibitor in the presence of benzamidine at 0.97 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-{2-[(4-carbamimidoylphenyl)carbamoyl]-6-methoxypyridin-3-yl}-5-{[(2S)-1-hydroxy-3,3-dimethylbutan-2-yl]carbamoyl}benzoic acid, BENZAMIDINE, ... | | Authors: | Sherawat, M, Kaur, P, Perbandt, M, Betzel, C, Slusarchyk, W.A, Bisacchi, G.S, Chang, C, Jacobson, B.L, Einspahr, H.M, Singh, T.P. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Structure of the complex of trypsin with a highly potent synthetic inhibitor at 0.97 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2AOS
 
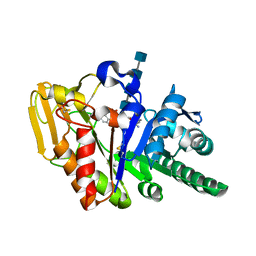 | | Protein-protein Interactions of protective signalling factor: Crystal structure of ternary complex involving signalling protein from goat (SPG-40), tetrasaccharide and a tripeptide Trp-pro-Trp at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Signaling protein from goat, SPG-40, ... | | Authors: | Kumar, J, Ethayathulla, A.S, Srivastava, D.B, Somvanshi, R.K, Singh, N, Sharma, S, Dey, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2005-08-14 | | Release date: | 2005-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein-protein Interactions of protective signalling factor: Crystal structure of ternary complex involving signalling protein from goat (SPG-40), tetrasaccharide and a tripeptide Trp-pro-Trp at 2.9 A resolution
To be Published
|
|
4QAJ
 
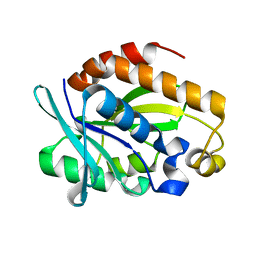 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Kumar, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2014-05-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
5YH7
 
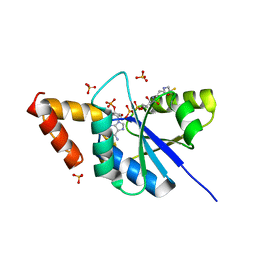 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Coenzyme A at 2.0 A resolution | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and binding studies of phosphopantetheine adenylyl transferase from Acinetobacter baumannii.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
5Z3S
 
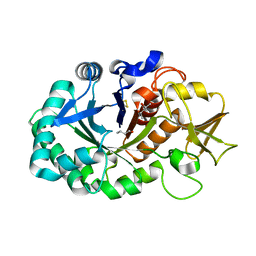 | | Crystal structure of butanol modified signaling protein from buffalo (SPB-40) at 1.65 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-BUTANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Chaudhary, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-01-08 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A glycoprotein from mammary gland secreted during involution promotes apoptosis: Structural and biological studies.
Arch. Biochem. Biophys., 644, 2018
|
|
