1YP7
 
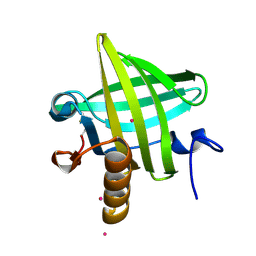 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | CADMIUM ION, MAJOR URINARY PROTEIN 1 | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1YP6
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1ZSE
 
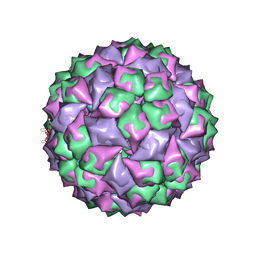 | | RNA stemloop from bacteriophage Qbeta complexed with an N87S mutant MS2 Capsid | | Descriptor: | Coat protein, RNA HAIRPIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-24 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2B2E
 
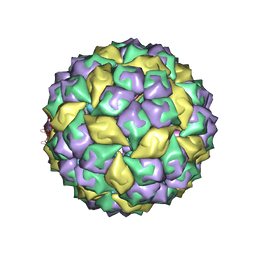 | | RNA stemloop from bacteriophage MS2 complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2B2D
 
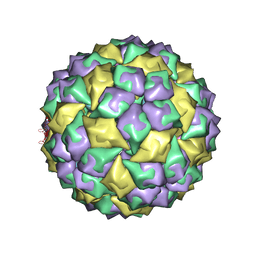 | | RNA stemloop operator from bacteriophage QBETA complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*UP*GP*CP*AP*UP*GP*UP*CP*UP*AP*AP*GP*AP*CP*AP*GP*CP*AP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2BS0
 
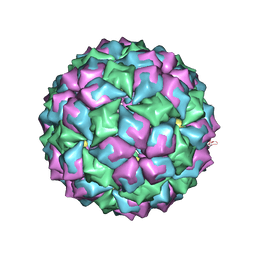 | | MS2 (N87AE89K mutant) - Variant Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*UP*GP*CP*AP*UP*GP*UP*CP*UP *AP*AP*GP*AP*CP*UP*GP*CP*AP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BS1
 
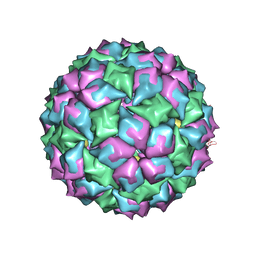 | | MS2 (N87AE89K mutant) - Qbeta RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-05-13 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BNY
 
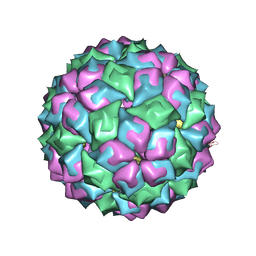 | | MS2 (N87A mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-06 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2BQ5
 
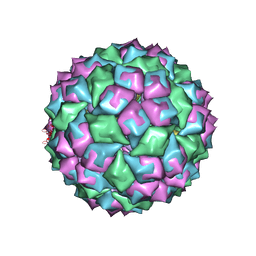 | | MS2 (N87AE89K mutant) - RNA hairpin complex | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *UP*AP*CP*CP*CP*AP*UP*GP*U)-3', COAT PROTEIN | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-04-27 | | Release date: | 2006-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Basis of RNA Binding Discrimination between Bacteriophages Qbeta and MS2.
Structure, 14, 2006
|
|
2B2G
 
 | | MS2 Wild-type RNA stemloop complexed with an N87S mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
1DYU
 
 | | The active site base controls cofactor reactivity in Escherichia coli amine oxidase: X-ray crystallographic studies with mutational variants. | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE | | Authors: | Murray, J.M, Wilmot, C.M, Saysell, C.G, Jaeger, J, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2000-02-08 | | Release date: | 2000-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The Active Site Base Controls Cofactor Reactivity in Escherichia Coli Amine Oxidase : X-Ray Crystallographicstudies with Mutational Variants
Biochemistry, 38, 1999
|
|
5KE0
 
 | | Discovery of 1-1H-Pyrazolo 4,3-c pyridine-6-yl urea Inhibitors of Extracellular Signal Regulated Kinase ERK for the Treatment of Cancers | | Descriptor: | 1-[3-(2-methylpyridin-4-yl)-1~{H}-pyrazolo[4,3-c]pyridin-6-yl]-3-(phenylmethyl)urea, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Hruza, A, Lim, J. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of 1-(1H-Pyrazolo[4,3-c]pyridin-6-yl)urea Inhibitors of Extracellular Signal-Regulated Kinase (ERK) for the Treatment of Cancers.
J.Med.Chem., 59, 2016
|
|
8FRR
 
 | | Wild-type myocilin olfactomedin domain | | Descriptor: | CALCIUM ION, GLYCEROL, Myocilin, ... | | Authors: | Ma, M.T, Lieberman, R.L, Huard, D.J.E. | | Deposit date: | 2023-01-08 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Competition between inside-out unfolding and pathogenic aggregation in an amyloid-forming beta-propeller.
Nat Commun, 15, 2024
|
|
4WXU
 
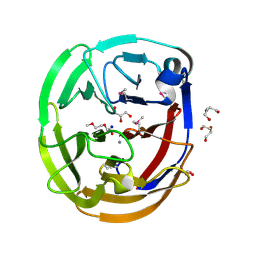 | | Crystal Structure of the Selenomthionine Incorporated Myocilin Olfactomedin Domain E396D Variant. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
4WXS
 
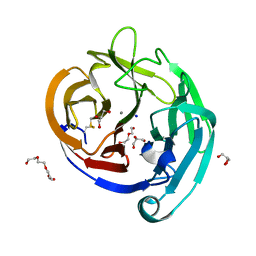 | | Crystal Structure of the E396D SNP Variant of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Donegan, R.K, Freeman, D.M, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
4WXQ
 
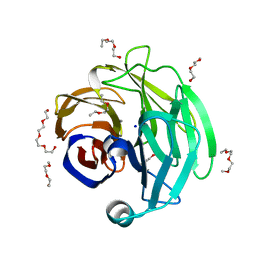 | | Crystal Structure of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Myocilin, ... | | Authors: | Orwig, S.D, Turnage, K.C, Donegan, R.K, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
6EG3
 
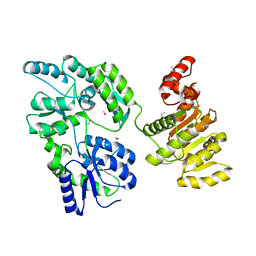 | | Crystal structure of human BRM in complex with compound 15 | | Descriptor: | 3-[(4-{[(2-chloropyridin-4-yl)carbamoyl]amino}pyridin-2-yl)ethynyl]benzoic acid, ETHANOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2 | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6EG2
 
 | | Crystal structure of human BRM in complex with compound 16 | | Descriptor: | ISOPROPYL ALCOHOL, Maltose/maltodextrin-binding periplasmic protein,Probable global transcription activator SNF2L2, N-(5-amino-2-chloropyridin-4-yl)-N'-(4-bromo-3-{[3-(hydroxymethyl)phenyl]ethynyl}-1,2-thiazol-5-yl)urea | | Authors: | Zhu, X, Kulathila, R, Hu, T, Xie, X. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers.
J. Med. Chem., 61, 2018
|
|
6CEQ
 
 | |
5A4M
 
 | | Mechanism of Hydrogen activation by NiFe-hydrogenases | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Evans, R.M, Brooke, E.J, Wehlin, S.A.M, Nomerotskaia, E, Sergent, F, Carr, S.B, Philips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
5EJT
 
 | | Thermally annealed ferryl Cytochrome C Peroxidase crystal structure | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EJX
 
 | | X-ray Free Electron Laser Structure of Cytochrome C Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PHOSPHATE ION, ... | | Authors: | Doukov, T, Soltis, S.M, Baxter, E.L, Cohen, A, Song, J, McPhillips, S, Poulos, T.L, Meharenna, Y.T, Chreifi, G. | | Deposit date: | 2015-11-02 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the pristine peroxidase ferryl center and its relevance to proton-coupled electron transfer.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LRY
 
 | | E coli [NiFe] Hydrogenase Hyd-1 mutant E28D | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J.Am.Chem.Soc., 140, 2018
|
|
6NNA
 
 | | Human Fatty Acid Synthase Psi/KR Tri-Domain with NADPH and Compound 22 | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid synthase,Fatty acid synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Toms, A.V, Martin, M.W. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery and optimization of novel piperazines as potent inhibitors of fatty acid synthase (FASN).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
6APK
 
 | | Trans-acting transferase from Disorazole synthase solved by serial femtosecond XFEL crystallography | | Descriptor: | DisD protein | | Authors: | Lyubimov, A.Y, Mathews, I.I, Uervivojnangkoorn, M, Khosla, C, Soltis, S.M, Cohen, A.E. | | Deposit date: | 2017-08-17 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Conformational Flexibility of the Acyltransferase from the Disorazole Polyketide Synthase Is Revealed by an X-ray Free-Electron Laser Using a Room-Temperature Sample Delivery Method for Serial Crystallography.
Biochemistry, 56, 2017
|
|
