1WV5
 
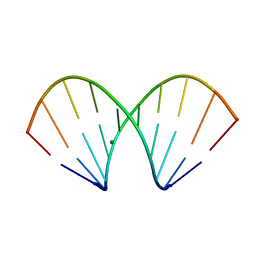 | | X-ray structure of the A-decamer GCGTATACGC with a single 2'-o-butyl thymine in place of T6, Mg-form | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2BT)P*AP*CP*GP*C)-3', MAGNESIUM ION | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharam, M. | | Deposit date: | 2004-12-11 | | Release date: | 2005-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2'-O-ribonucleic acid modifications.
Biochemistry, 44, 2005
|
|
3IR2
 
 | |
7AC0
 
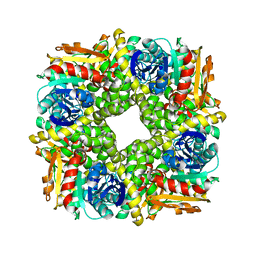 | | Epoxide hydrolase CorEH without ligand | | Descriptor: | Soluble epoxide hydrolase | | Authors: | Palm, G.J, Lammers, M, Berndt, L. | | Deposit date: | 2020-09-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Promiscuous Dehalogenase Activity of the Epoxide Hydrolase CorEH from Corynebacterium sp. C12
Acs Catalysis, 11, 2021
|
|
6ST8
 
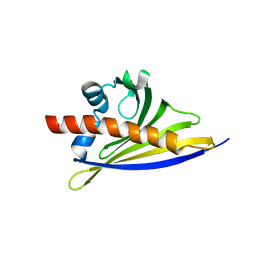 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
6ST9
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, D48R mutant | | Descriptor: | CHLORIDE ION, Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
6STA
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, E46A D48A mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
468D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
470D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
469D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
7LO9
 
 | | RNA dodecamer containing a GNA A residue | | Descriptor: | Chains: A,B,C,D | | Authors: | Harp, J.M, Wawrzak, Z, Egli, M. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs.
Nucleic Acids Res., 49, 2021
|
|
6VEM
 
 | | Structure of RNA octamer | | Descriptor: | COBALT HEXAMMINE(III), Modified Octamer RNA | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Synthesis, chirality-dependent conformational and biological properties of siRNAs containing 5'-(R)- and 5'-(S)-C-methyl-guanosine.
Nucleic Acids Res., 48, 2020
|
|
6STB
 
 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, Q64W mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
471D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
2JN0
 
 | | Solution NMR structure of the ygdR protein from Escherichia coli. Northeast Structural Genomics target ER382A. | | Descriptor: | Hypothetical lipoprotein ygdR | | Authors: | Rossi, P, Chen, C.X, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J.C, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-15 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the ygdR protein from Escherichia coli. Northeast Structural Genomics target ER382A.
To be Published
|
|
2HEP
 
 | | Solution NMR structure of the UPF0291 protein ynzC from Bacillus subtilis. Northeast Structural Genomics target SR384. | | Descriptor: | UPF0291 protein ynzC | | Authors: | Aramini, J.M, Swapna, G.V.T, Ho, C.K, Shetty, K, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the SOS response protein YnzC from Bacillus subtilis
Proteins, 72, 2008
|
|
8USR
 
 | | IL17A homodimer complexed to Compound 23 | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1-[[(1~{S})-1-(8~{a}~{H}-imidazo[1,2-a]pyrimidin-2-yl)ethyl]amino]-1-oxidanylidene-4-phenyl-butan-2-yl]-4,5-bis(chloranyl)-1~{H}-pyrrole-2-carboxamide | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
8USS
 
 | | IL17A complexed to Compound 7 | | Descriptor: | 4,5-dichloro-N-[(1S)-1-cyclohexyl-2-{[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]amino}-2-oxoethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, Interleukin-17A | | Authors: | Argiriadi, M.A, Ramos, A.L. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of Small Molecule Interleukin 17A Inhibitors with Novel Binding Mode and Stoichiometry: Optimization of DNA-Encoded Chemical Library Hits to In Vivo Active Compounds.
J.Med.Chem., 67, 2024
|
|
6O6J
 
 | |
6O7A
 
 | |
6O7C
 
 | |
3EY3
 
 | |
2INW
 
 | | Crystal structure of Q83JN9 from Shigella flexneri at high resolution. Northeast Structural Genomics Consortium target SfR137. | | Descriptor: | PHOSPHATE ION, Putative structural protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Vorobiev, S.M, Wang, D, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-09 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3EY2
 
 | |
3EY1
 
 | |
2FIH
 
 | |
