7DCA
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase bound by xanthosine | | Descriptor: | 2,3-dihydroxanthosine, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-10-23 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The C-terminal loop of Arabidopsis thaliana guanosine deaminase is essential to catalysis.
Chem.Commun.(Camb.), 57, 2021
|
|
7DBF
 
 | |
7DC9
 
 | |
7E8L
 
 | | The structure of Spodoptera litura chemosensory protein | | Descriptor: | Putative chemosensory protein CSP8 | | Authors: | Xie, W, Jia, Q, Zeng, H, Xiao, N, Tang, J, Gao, S, Zhang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Spodoptera litura Chemosensory Protein CSP8.
Insects, 12, 2021
|
|
7DOX
 
 | |
7DOY
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in complex with 6-O-methylguanosine | | Descriptor: | (2R,3R,4S,5R)-2-(2-azanyl-6-methoxy-purin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DLC
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with N1-methylguanosine | | Descriptor: | 2-azanyl-9-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1-methyl-purin-6-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7DPK
 
 | |
7DOW
 
 | | The structure of the Arabidopsis thaliana guanosine deaminase in reaction with 7-deazaguansoine | | Descriptor: | 2-azanyl-7-[(2R,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-3H-pyrrolo[2,3-d]pyrimidin-4-one, Guanosine deaminase, ZINC ION | | Authors: | Xie, W, Jia, Q, Zeng, H. | | Deposit date: | 2020-12-17 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric Catalysis of Arabidopsis thaliana Guanosine Deaminase Revealed by Crystal Structures
To Be Published
|
|
7WR1
 
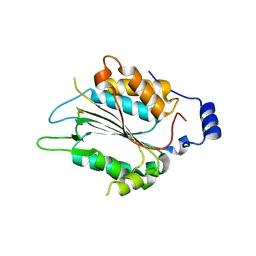 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | Descriptor: | Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR4
 
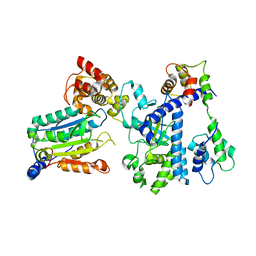 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR0
 
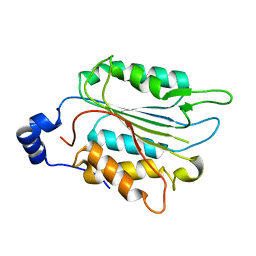 | | P32 of caspase-4 C258A mutant | | Descriptor: | Caspase-4 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR6
 
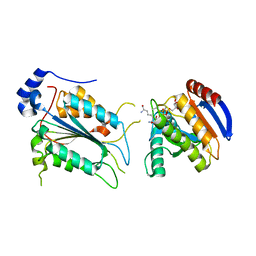 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
 | |
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2RFI
 
 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and dimethylated H3K9 peptide | | Descriptor: | Histone H3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
5FID
 
 | |
5JJZ
 
 | | Chromo domain of human Chromodomain Protein, Y-Like 2 | | Descriptor: | Chromodomain Y-like protein 2, LYS-LYS-LYS-ALA-ARG-MLY-SER-ALA-GLY-ALA-ALA-LYS-TYR | | Authors: | DONG, A, DOMBROVSKI, L, LOPPNAU, P, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, MIN, J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-04-25 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of CDYL2 domain of human CDYL2 protein
to be published
|
|
2TDX
 
 | | DIPHTHERIA TOX REPRESSOR (C102D MUTANT) COMPLEXED WITH NICKEL | | Descriptor: | DIPHTHERIA TOX REPRESSOR, NICKEL (II) ION | | Authors: | White, A, Ding, X, Zheng, H, Schiering, N, Ringe, D, Murphy, J.R. | | Deposit date: | 1998-06-22 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex.
Nature, 394, 1998
|
|
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
3HNA
 
 | | Crystal structure of catalytic domain of human euchromatic histone methyltransferase 1 in complex with SAH and mono-Methylated H3K9 Peptide | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, Mono-Methylated H3K9 Peptide, ... | | Authors: | Min, J, Wu, H, Loppnau, P, Wleigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural biology of human H3K9 methyltransferases
Plos One, 5, 2010
|
|
8TU6
 
 | | CryoEM structure of PI3Kalpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Valverde, R, Shi, H, Holliday, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TSA
 
 | | Human PI3K p85alpha/p110alpha H1047R bound to compound 2 | | Descriptor: | 5-(3-bromo-5-fluorobenzamido)-N-methyl-6-(2-methylanilino)pyridine-3-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J, Valverde, R. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
8TS8
 
 | | p85alpha/p110alpha heterodimer H1047R mutant | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Holliday, M, Tang, Y, Bulku, A, Wilbur, J, Fraser, J. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
