8QC8
 
 | |
8QC2
 
 | | Crystal structure of NAD-dependent glycoside hydrolase from Flavobacterium sp. (strain K172) in complex with co-factor NAD+ and sulfoquinovose (SQ) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Pickles, I.B, Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Widespread Family of NAD + -Dependent Sulfoquinovosidases at the Gateway to Sulfoquinovose Catabolism.
J.Am.Chem.Soc., 145, 2023
|
|
8QC3
 
 | |
8R56
 
 | |
8P0L
 
 | | Crystal structure of human O-GlcNAcase in complex with an S-linked CKII peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Protein O-GlcNAcase, ... | | Authors: | Males, A, Davies, G.J, Calvelo, M, Alteen, M.G, Vocadlo, D.J, Rovira, C. | | Deposit date: | 2023-05-10 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human O -GlcNAcase Uses a Preactivated Boat-skew Substrate Conformation for Catalysis. Evidence from X-ray Crystallography and QM/MM Metadynamics.
Acs Catalysis, 13, 2023
|
|
8QC5
 
 | |
8QC6
 
 | |
8R1M
 
 | | Structure of TxGH116 with covalently bound N-azido-octyl aziridine | | Descriptor: | (1R,2R,3R,4S)-4-[8-(azanyldiazenyl)octylamino]-3-(hydroxymethyl)cyclopentane-1,2-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective labelling of GBA2 in cells with fluorescent beta-d-arabinofuranosyl cyclitol aziridines.
Chem Sci, 15, 2024
|
|
8OHX
 
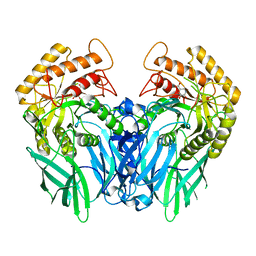 | | Crystal structure of Beta-glucuronidase from Escherichia coli in complex with siastatin B derived inhibitor | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, Beta-D-glucuronidase | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OGX
 
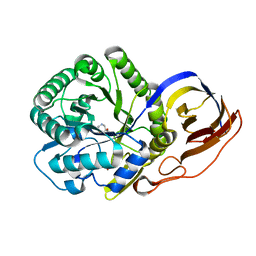 | | Beta-glucuronidase from Acidobacterium capsulatum in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, PHOSPHATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHT
 
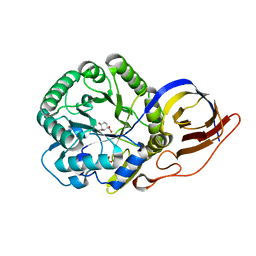 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHU
 
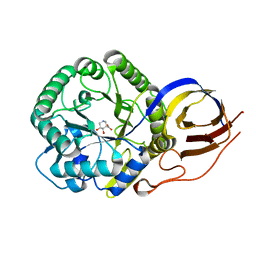 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured isofagamine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHQ
 
 | | Crystal structure of human heparanase in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Davies, G.J, Overkleeft, H.S, Armstrong, Z, Yurong, C. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHV
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured 3-geminal diol iminosugar inhibitor | | Descriptor: | (3~{S},4~{R})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHR
 
 | | Crystal structure of human heparanase in complex with glucuronic acid configured 3-geminal diol iminosugar inhibitor | | Descriptor: | (3~{S},4~{R})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHW
 
 | | Crystal structure of heparanase from Burkholderia pseudomallei in complex with siastatin B derived inhibitor | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, Glycoside hydrolase family 44 domain-containing protein | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
7OU6
 
 | | Human O-GlcNAc hydrolase in complex with DNJNAc-thiazolidines | | Descriptor: | Protein O-GlcNAcase, ~{N}-[(3~{Z},6~{S},7~{R},8~{R},8~{a}~{S})-7,8-bis(oxidanyl)-3-(phenylmethyl)imino-1,5,6,7,8,8~{a}-hexahydro-[1,3]thiazolo[3,4-a]pyridin-6-yl]ethanamide | | Authors: | Males, A, Davies, G.J, Gonzalez-Cuesta, M, Mellet, C.O, Fernandez, J.M.G, Sidhu, P, Ashmus, R, Busmann, J, Vocadlo, D.J, Foster, L. | | Deposit date: | 2021-06-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Bicyclic Picomolar OGA Inhibitors Enable Chemoproteomic Mapping of Its Endogenous Post-translational Modifications
J.Am.Chem.Soc., 144, 2022
|
|
7OP6
 
 | | Bacteroides thetaiotaomicron mannosidase GH2 with beta-manno-configured cyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, BROMIDE ION, ... | | Authors: | McGregor, N.G.S, Beenakker, T.J.M, Kuo, C, Wong, C, Offen, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
4MAI
 
 | | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) | | Descriptor: | 1,2-ETHANEDIOL, AA11 Lytic Polysaccharide Monooxygenase, CHLORIDE ION, ... | | Authors: | Hemsworth, G.R, Henrissat, B, Walton, P.H, Davies, G.J. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and characterization of a new family of lytic polysaccharide monooxygenases.
Nat.Chem.Biol., 10, 2014
|
|
5C86
 
 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, apo form | | Descriptor: | Kelch domain-containing protein | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
5C92
 
 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, in complex with copper | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (I) ION, ... | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
4MAH
 
 | | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn | | Descriptor: | 1,2-ETHANEDIOL, AA11 Lytic Polysaccharide Monooxygenase, CHLORIDE ION, ... | | Authors: | Hemsworth, G.R, Henrissat, B, Walton, P.H, Davies, G.J. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-18 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a new family of lytic polysaccharide monooxygenases.
Nat.Chem.Biol., 10, 2014
|
|
4OPB
 
 | | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Frandsen, K.H, Davies, G.J, Dupree, P, Walton, P, Henrissat, B, Stringer, M, Tovborg, M, De Maria, L, Johansen, K.S. | | Deposit date: | 2014-02-05 | | Release date: | 2015-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase.
Nat Commun, 6, 2015
|
|
7QHV
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with sulfoquinovosyl diacylglycerol | | Descriptor: | GLYCINE, Sulfoquinovosyl binding protein, [(2~{S},3~{S},4~{S},5~{R},6~{S})-6-[(2~{S})-3-butanoyloxy-2-heptanoyloxy-propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A, Davies, G.J. | | Deposit date: | 2021-12-14 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7QQH
 
 | | Crystal structure of MYORG (D520N) in complex with Gal-a1,4-Glc | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The primary familial brain calcification-associated protein MYORG is an alpha-galactosidase with restricted substrate specificity.
Plos Biol., 20, 2022
|
|
