7Y3L
 
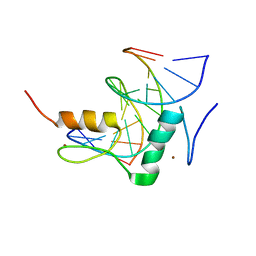 | | Structure of SALL3 ZFC4 bound with 12 bp AT-rich dsDNA | | Descriptor: | DNA (12-mer), Sal-like protein 3, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7Y3M
 
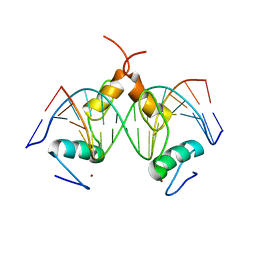 | | Structure of SALL4 ZFC1 bound with 16 bp AT-rich dsDNA | | Descriptor: | DNA (16-mer), Sal-like protein 4, ZINC ION | | Authors: | Ru, W, Xu, C. | | Deposit date: | 2022-06-11 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.723 Å) | | Cite: | Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif.
J.Biol.Chem., 298, 2022
|
|
7EBJ
 
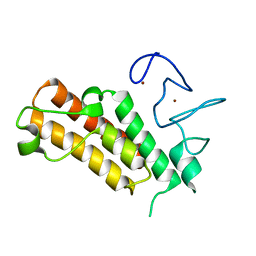 | | Apo structure of the mouse Trim66 PHD-Bromo dual domain | | Descriptor: | Tripartite motif-containing protein 66, ZINC ION | | Authors: | Wang, Z, Jiang, J. | | Deposit date: | 2021-03-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A TRIM66/DAX1/Dux axis suppresses the totipotent 2-cell-like state in murine embryonic stem cells.
Cell Stem Cell, 29, 2022
|
|
8IMW
 
 | |
8J0D
 
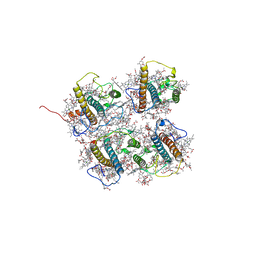 | | FCP heterodimer, Lhca2, and Lhcf5 together as the M1 side binds to the PSII core in the diatom Thalassiosira pseudonana | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z, Feng, Y, Wang, W, Shen, J.R. | | Deposit date: | 2023-04-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
8IWH
 
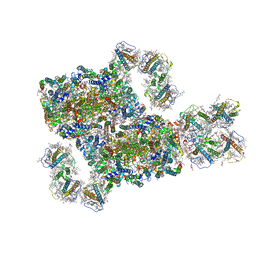 | | Structure and characteristics of a photosystem II supercomplex containing monomeric LHCX and dimeric FCPII antennae from the diatom Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z.H, Wang, W.D, Shen, J.R. | | Deposit date: | 2023-03-30 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
8IVZ
 
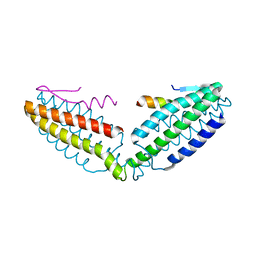 | | Crystal structure of talin R7 in complex with KANK1 KN motif | | Descriptor: | KN motif and ankyrin repeat domains 1, Talin-1 | | Authors: | Xu, Y, Li, K, Wei, Z, Cong, Y. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
7XGW
 
 | |
8J17
 
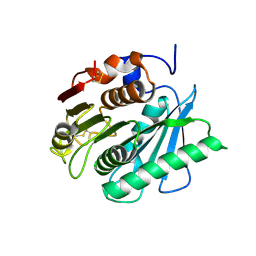 | | Crystal structure of IsPETase variant | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Yin, Q.D, Wang, Y.X. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Efficient polyethylene terephthalate biodegradation by an engineered Ideonella sakaiensis PETase with a fixed substrate-binding W156 residue
Green Chem, 2013
|
|
8I3G
 
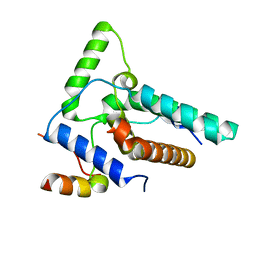 | | Crystal structure of Eaf3-Eaf7 complex | | Descriptor: | Chromatin modification-related protein EAF3, Chromatin modification-related protein EAF7 | | Authors: | Chen, Z, Xu, C. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for Eaf3-mediated assembly of Rpd3S and NuA4.
Cell Discov, 9, 2023
|
|
8I3F
 
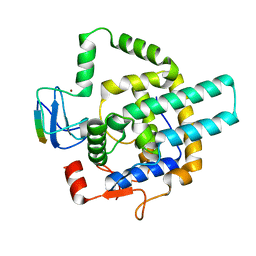 | |
7YIM
 
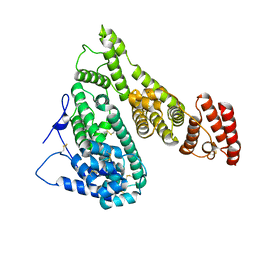 | | Cryo-EM structure of human Alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein | | Authors: | Liu, N, Liu, K, Wu, C, Liu, Z, Li, M, Wang, J, Wang, H.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
7FFT
 
 | |
7FFW
 
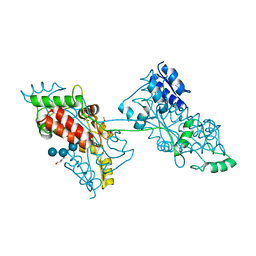 | | The crystal structure of a domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, METHOXYETHANE, ... | | Authors: | Wang, L, Bu, T, Bai, X. | | Deposit date: | 2021-07-23 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the domain-swapped dimeric maltodextrin-binding protein MalE from Salmonella enterica.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7F1E
 
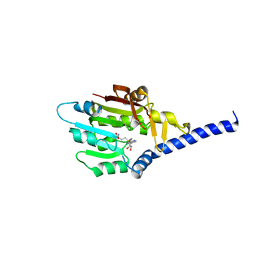 | | Structure of METTL6 bound with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Li, S, Liao, S, Xu, C. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Structural basis for METTL6-mediated m3C RNA methylation.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
7WIN
 
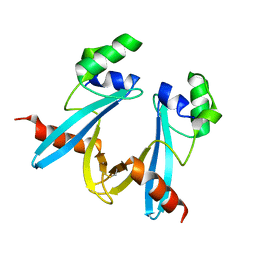 | |
7FJ1
 
 | | Cryo-EM structure of pseudorabies virus C-capsid | | Descriptor: | Capsid vertex component 1, DNA packaging tegument protein UL25, Major capsid protein, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
7FJ3
 
 | | Cryo-EM structure of PRV A-capid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
7W1Q
 
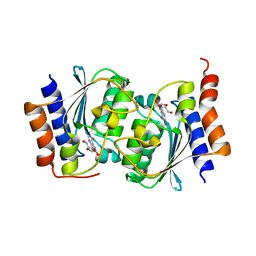 | |
7X39
 
 | | Structure of CIZ1 bound ERH | | Descriptor: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
7XB4
 
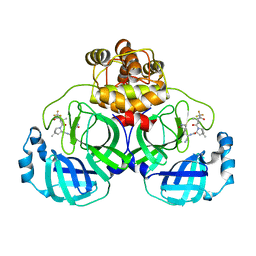 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Hu, X.H, Li, J, Zhang, J. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
7XGY
 
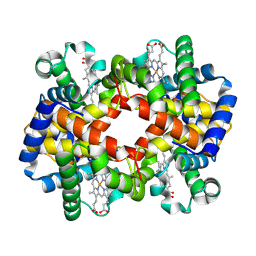 | | cryo-EM structure of hemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, N, Wang, H.W. | | Deposit date: | 2022-04-07 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
7VV9
 
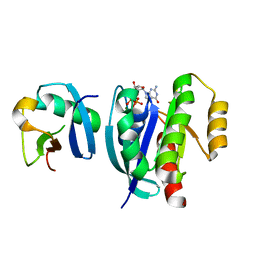 | |
7VVG
 
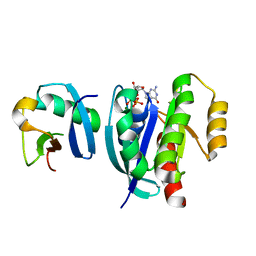 | |
7VVB
 
 | |
