4A0G
 
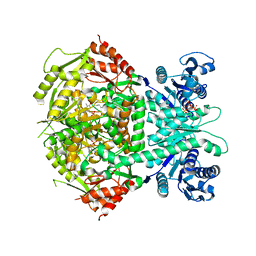 | | Structure of bifunctional DAPA aminotransferase-DTB synthetase from Arabidopsis thaliana in its apo form. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cobessi, D, Dumas, R, Pautre, V, Meinguet, C, Ferrer, J.L, Alban, C. | | Deposit date: | 2011-09-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Biochemical and Structural Characterization of the Arabidopsis Bifunctional Enzyme Dethiobiotin Synthetase-Diaminopelargonic Acid Aminotransferase: Evidence for Substrate Channeling in Biotin Synthesis.
Plant Cell, 24, 2012
|
|
3HQF
 
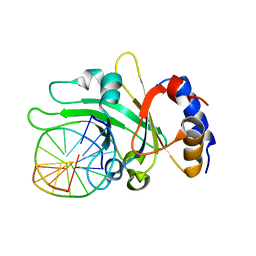 | | Crystal structure of restriction endonuclease EcoRII N-terminal effector-binding domain in complex with cognate DNA | | Descriptor: | 5'-D(*CP*GP*CP*CP*AP*GP*GP*GP*C)-3', 5'-D(*GP*CP*CP*CP*TP*GP*GP*CP*G)-3', Restriction endonuclease | | Authors: | Golovenko, D, Manakova, E, Grazulis, S, Tamulaitiene, G, Siksnys, V. | | Deposit date: | 2009-06-06 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII.
Nucleic Acids Res., 37, 2009
|
|
1CLU
 
 | | H-RAS COMPLEXED WITH DIAMINOBENZOPHENONE-BETA,GAMMA-IMIDO-GTP | | Descriptor: | 3-AMINOBENZOPHENONE-4-YL-AMINOHYDROXYPHOSPHINYLAMINOPHOSPHONIC ACID-GUANYLATE ESTER, MAGNESIUM ION, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Ahmadian, M.R, Zor, T, Vogt, D, Kabsch, W, Selinger, Z, Wittinghofer, A, Scheffzek, K. | | Deposit date: | 1999-05-03 | | Release date: | 1999-05-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanosine triphosphatase stimulation of oncogenic Ras mutants.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
424D
 
 | | 5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3' | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*T)-3') | | Authors: | Rozenberg, H, Rabinovich, D, Frolow, F, Hegde, R.S, Shakked, Z. | | Deposit date: | 1998-09-14 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2OKB
 
 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-16 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3FZ8
 
 | |
3JV4
 
 | |
4AAS
 
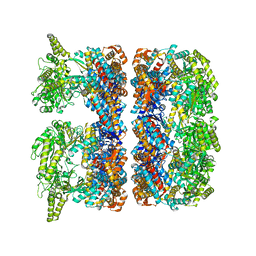 | | ATP-triggered molecular mechanics of the chaperonin GroEL | | Descriptor: | 60 KDA CHAPERONIN, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Clare, D.K, Vasishtan, D, Stagg, S, Quispe, J, Farr, G.W, Topf, M, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | ATP-Triggered Conformational Changes Delineate Substrate-Binding and -Folding Mechanics of the Groel Chaperonin.
Cell(Cambridge,Mass.), 149, 2012
|
|
6THF
 
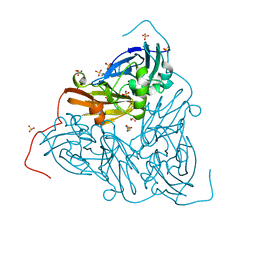 | | Crystal structure of two-domain Cu nitrite reductase from Bradyrhizobium sp. ORS 375 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-20 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Reverse protein engineering of a novel 4-domain copper nitrite reductase reveals functional regulation by protein-protein interaction.
Febs J., 288, 2021
|
|
6T1I
 
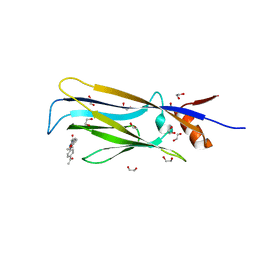 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-ethanoylphenyl)-~{N}-[(6-methoxypyridin-3-yl)methyl]piperazine-1-carboxamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
6T1O
 
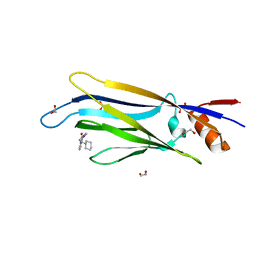 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
1D4A
 
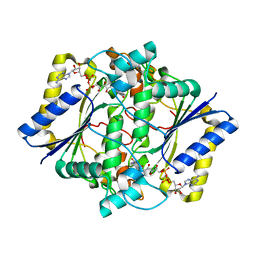 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE AT 1.7 A RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3X1I
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxo-tetracosahexaenoic acid | | Descriptor: | (8E,12Z,15Z,18Z,21Z)-6-oxotetracosa-8,12,15,18,21-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2014-11-18 | | Release date: | 2015-04-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of covalent bond formation between PPAR gamma and oxo-fatty acids.
Bioconjug.Chem., 26, 2015
|
|
6T6B
 
 | | Crystal structure of PPARgamma in complex with compound 16 (MF27) | | Descriptor: | (2~{R})-2-[[6-[(2,4-dichlorophenyl)sulfonylamino]-1,3-benzothiazol-2-yl]sulfanyl]octanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Ni, X, Hanke, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Selective Modulator of Peroxisome Proliferator-Activated Receptor gamma with an Unprecedented Binding Mode.
J.Med.Chem., 63, 2020
|
|
2OL1
 
 | | High Resolution Crystal Structures of Vaccinia Virus dUTPase | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Schormann, N, Chattopadhyay, D. | | Deposit date: | 2007-01-18 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of vaccinia virus dUTPase and its nucleotide complexes.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OMM
 
 | |
3JCG
 
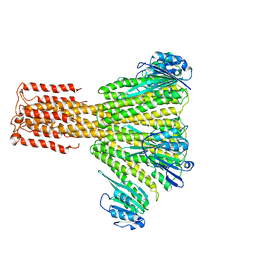 | |
3JD2
 
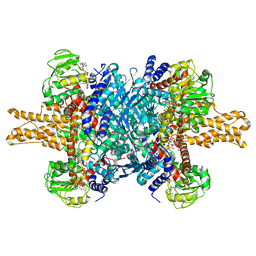 | | Glutamate dehydrogenase in complex with NADH, open conformation | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3G7W
 
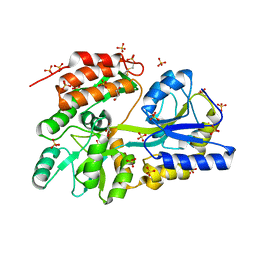 | | Islet Amyloid Polypeptide (IAPP or Amylin) Residues 1 to 22 fused to Maltose Binding Protein | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein, Islet amyloid polypeptide fusion protein, ... | | Authors: | Wiltzius, J.J.W, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2009-02-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process
Protein Sci., 18, 2009
|
|
2ON3
 
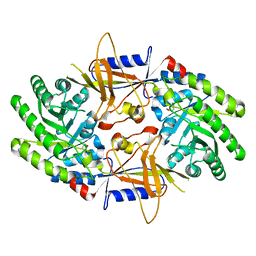 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase | | Authors: | Dufe, V.T, Ingner, D, Heby, O, Khomutov, A.R, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
3JRX
 
 | | Crystal structure of the BC domain of ACC2 in complex with soraphen A | | Descriptor: | Acetyl-CoA carboxylase 2, SORAPHEN A | | Authors: | Cho, Y.S, Lee, J.I, Shin, D, Kim, H.T, Lee, T.G, Heo, Y.S. | | Deposit date: | 2009-09-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism for the regulation of human ACC2 through phosphorylation by AMPK.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3FSH
 
 | |
3FSS
 
 | | Structure of the tandem PH domains of Rtt106 | | Descriptor: | GLYCEROL, Histone chaperone RTT106, MALONIC ACID | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2009-01-11 | | Release date: | 2009-12-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
6TA0
 
 | | Human NAMPT in complex with nicotinic acid and phosphoribosyl pyrophosphate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, NICOTINIC ACID, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
3K2G
 
 | | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, MAGNESIUM ION, Resiniferatoxin-binding, ... | | Authors: | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a Resiniferatoxin-binding protein from Rhodobacter sphaeroides
To be Published
|
|
