8WHU
 
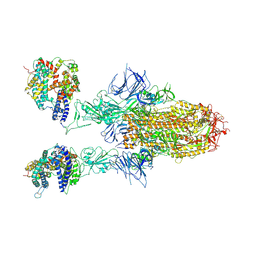 | | Spike Trimer of BA.2.86 in complex with two hACE2s | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5R
 
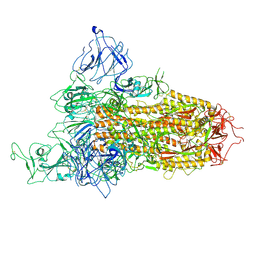 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHW
 
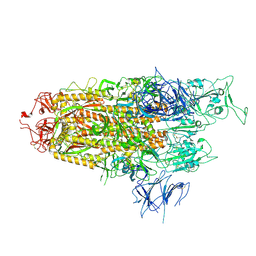 | | Spike Trimer of BA.2.86 with single RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUR
 
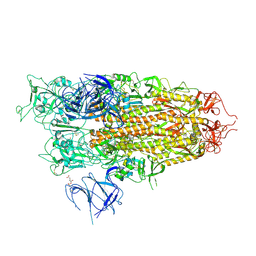 | | BA.2.86 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUU
 
 | | BA.2.86-T356K Spike Trimer in complex with heparan sulfate (Local refinement) | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8CXB
 
 | | Human PA28-20S (PA28-4a3b) | | Descriptor: | Proteasome activator complex subunit 1, Proteasome activator complex subunit 2, Proteasome subunit alpha type-1, ... | | Authors: | Zhao, J, Makhija, S, Huang, B, Cheng, Y. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8X50
 
 | | BA.2.86 Spike Trimer with ins483V mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4H
 
 | | SARS-CoV-2 JN.1 Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5Q
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUT
 
 | | XBB.1.5 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P, Mao, X. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHV
 
 | | Spike Trimer of BA.2.86 with three RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHS
 
 | | Spike Trimer of BA.2.86 in complex with one hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
7YS6
 
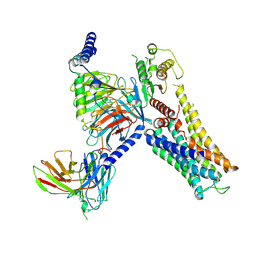 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5WJQ
 
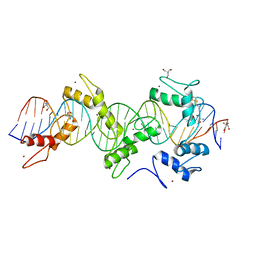 | | mouseZFP568-ZnF2-11 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (28-MER), ZINC ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
5JHP
 
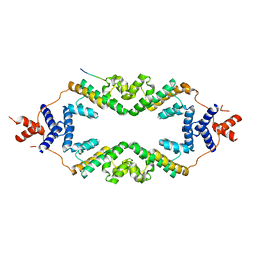 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L179A and I195A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 EAR peptide (794-808) | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
7YFM
 
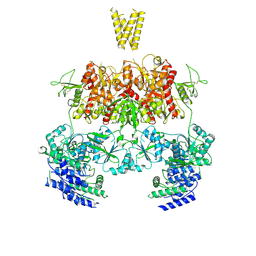 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFF
 
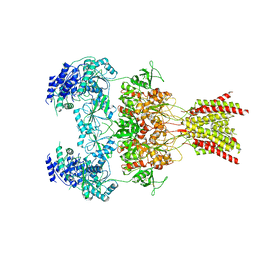 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
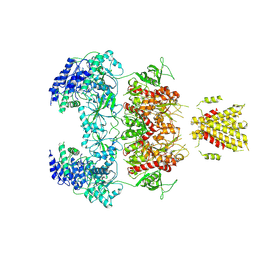 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
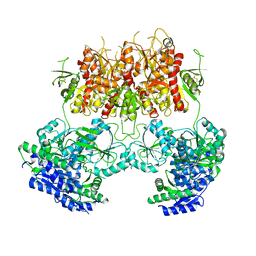 | |
7YFR
 
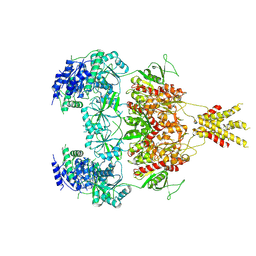 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, J.L, Zhu, S.J, Zhang, M. | | Deposit date: | 2022-07-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5JGC
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A, L130A, L179A and I195A mutant | | Descriptor: | Protein TPR1, ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-20 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5H25
 
 | | EED in complex with PRC2 allosteric inhibitor compound 11 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
5H24
 
 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
6LS5
 
 | | Structure of human liver FBPase complexed with covalent allosteric inhibitor | | Descriptor: | 2-(ethyldisulfanyl)-1,3-benzothiazole, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Yunyuan, H, Rongrong, S, Yixiang, X, Shuaishuai, N, Yanliang, R, Jian, L, Jian, W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Identification of the New Covalent Allosteric Binding Site of Fructose-1,6-bisphosphatase with Disulfiram Derivatives toward Glucose Reduction.
J.Med.Chem., 63, 2020
|
|
