6EXF
 
 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Fe(II)/Lysine | | Descriptor: | FE (III) ION, GLYCEROL, L-lysine 4-hydroxylase, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
6F9P
 
 | | Crystal structure of the Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Re(II) | | Descriptor: | CHLORIDE ION, GLYCEROL, L-lysine 4-hydroxylase, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
6EUO
 
 | | Crystal structure of APO Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 | | Descriptor: | D-MALATE, FE (III) ION, GLYCEROL, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-10-30 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
6EXH
 
 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Fe(II)/succinate/(4R)-4-hydroxy-L-lysine | | Descriptor: | 4-HYDROXY-LYSINE, FE (III) ION, GLYCEROL, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
6FI9
 
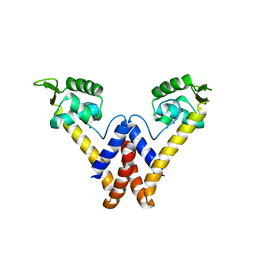 | |
6EUR
 
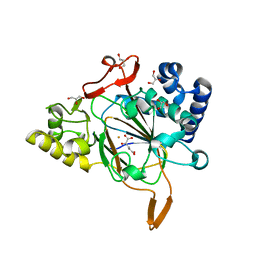 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Fe(II)/alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
6F2E
 
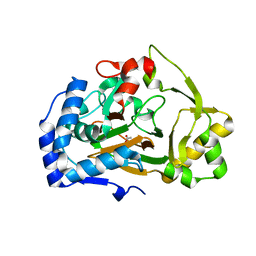 | | Crystal structure of the APO Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO1 | | Descriptor: | ACETIC ACID, L-lysine 3-hydroxylase | | Authors: | Isabet, T, Stura, E.A, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-11-24 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
6F6J
 
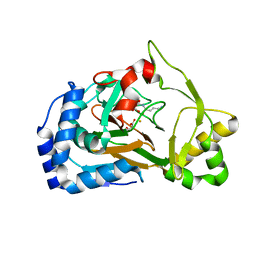 | | Crystal structure of the Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO1 with Fe(II)/succinate/(3S)-3-hydroxy-L-lysine | | Descriptor: | (2~{S},3~{R})-2,6-bis(azanyl)-3-oxidanyl-hexanoic acid, ACETATE ION, FE (III) ION, ... | | Authors: | Isabet, T, Stura, E.A, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-12-05 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
7BDX
 
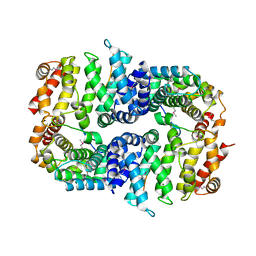 | | Armadillo domain of HSF2BP in complex with BRCA2 peptide | | Descriptor: | Breast cancer type 2 susceptibility protein, Heat shock factor 2-binding protein, MAGNESIUM ION | | Authors: | Le Du, M.H, Zinn-Justin, S, Ghouil, R, Miron, S, Legrand, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | BRCA2 binding through a cryptic repeated motif to HSF2BP oligomers does not impact meiotic recombination.
Nat Commun, 12, 2021
|
|
7ANQ
 
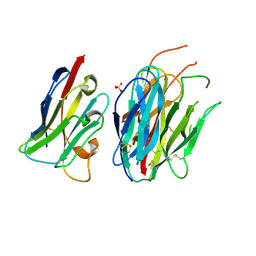 | | Complete PCSK9 C-ter domain in complex with VHH P1.40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Proprotein convertase subtilisin/kexin type 9, SULFATE ION, ... | | Authors: | Ciccone, L, Legrand, P, Stura, E.A, Dive, V, Seidahn, N.G, Fruchart Gaillard, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular interactions of PCSK9 with an inhibitory nanobody, CAP1 and HLA-C: Functional regulation of LDLR levels.
Mol Metab, 67, 2022
|
|
7AWP
 
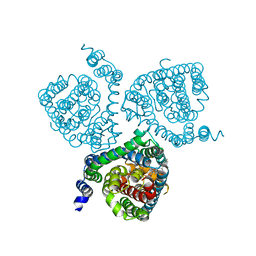 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with rubidium and barium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWM
 
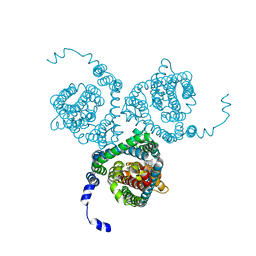 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with L-ASP, three sodium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, BARIUM ION, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWL
 
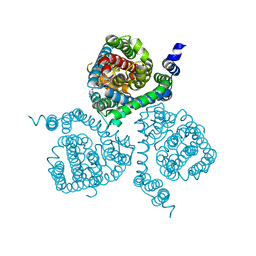 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with barium and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWQ
 
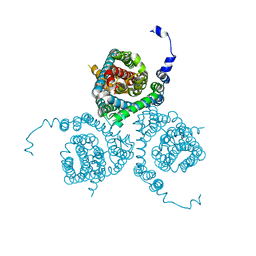 | | Structure of the thermostabilized EAAT1 cryst-E386Q mutant in complex with L-ASP, sodium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, BARIUM ION, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWN
 
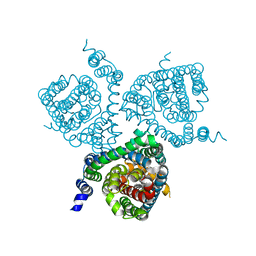 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with rubidium and barium and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
6SCY
 
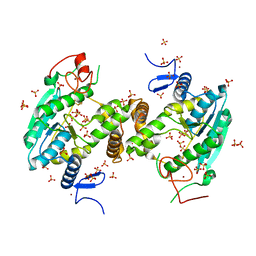 | | U34-tRNA thiolase NcsA from Methanococcus maripaludis with its [4Fe-4S] cluster | | Descriptor: | IRON/SULFUR CLUSTER, SULFATE ION, ZINC ION, ... | | Authors: | Bimai, O, Legrand, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2019-07-25 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The thiolation of uridine 34 in tRNA, which controls protein translation, depends on a [4Fe-4S] cluster in the archaeum Methanococcus maripaludis.
Sci Rep, 13, 2023
|
|
6TMR
 
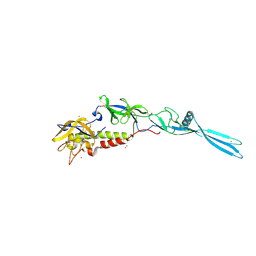 | | Mokola virus glycoprotein, monomeric post-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, TERBIUM(III) ION | | Authors: | Belot, L, Roche, S, Legrand, P, Gaudin, Y, Albertini, A. | | Deposit date: | 2019-12-05 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Crystal structure of Mokola virus glycoprotein in its post-fusion conformation.
Plos Pathog., 16, 2020
|
|
6T1Z
 
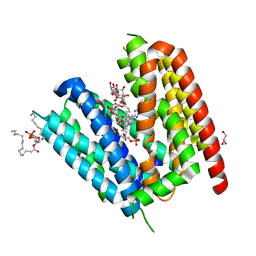 | | LmrP from L. lactis, in an outward-open conformation, bound to Hoechst 33342 | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DODECYL-ALPHA-D-MALTOSIDE, ... | | Authors: | Hutchin, A, Debruycker, V, Masureel, M, Legrand, P, Remaut, H, Govaerts, C. | | Deposit date: | 2019-10-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An embedded lipid in the multidrug transporter LmrP suggests a mechanism for polyspecificity.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8A51
 
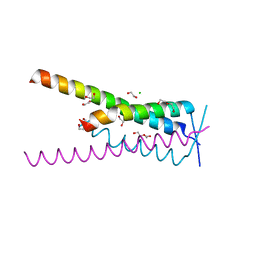 | | Crystal structure of HSF2BP-BRME1 complex | | Descriptor: | 1,2-ETHANEDIOL, Break repair meiotic recombinase recruitment factor 1, CHLORIDE ION, ... | | Authors: | Miron, S, Legrand, P, Ropars, V, Ghouil, R, Zinn-Justin, S. | | Deposit date: | 2022-06-13 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BRCA2-HSF2BP oligomeric ring disassembly by BRME1 promotes homologous recombination.
Sci Adv, 9, 2023
|
|
8AI6
 
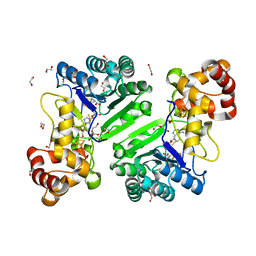 | | Crystal structure of radical SAM epimerase EpeE D210A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and persulfurated cysteine bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI1
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-homocysteine bound. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Polsinelli, I, Chavas, L.M.G, Legrand, P, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI4
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI3
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters and S-adenosyl-L-methionine bound | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kubiak, X, Chavas, L.M.G, Legrand, P, Polsinelli, I, Fyfe, C.D, Benjdia, A, Berteau, O. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI2
 
 | | Crystal structure of radical SAM epimerase EpeE from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 5 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
8AI5
 
 | | Crystal structure of radical SAM epimerase EpeE C223A mutant from Bacillus subtilis with [4Fe-4S] clusters, S-adenosyl-L-homocysteine and RiPP peptide 6 bound | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IRON/SULFUR CLUSTER, ... | | Authors: | Polsinelli, I, Fyfe, C.D, Legrand, P, Kubiak, X, Chavas, L.M.G, Berteau, O, Benjdia, A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Nat.Chem.Biol., 20, 2024
|
|
