5F86
 
 | |
8TQS
 
 | |
4DZO
 
 | |
6P6G
 
 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6P6K
 
 | |
4MJ5
 
 | | Crystal Structure of HLA-A*1101 in complex with H1-22, an influenza A(H1N1) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
4MJ6
 
 | | Crystal Structure of HLA-A*1101 in complex with H7-22, an influenza A(H7N9) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
5JA5
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
5JHP
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L179A and I195A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 EAR peptide (794-808) | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
6VLC
 
 | |
6VU6
 
 | |
6VS7
 
 | | Sialic acid binding region of Streptococcus Sanguinis SK1 adhesin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adhesin, ... | | Authors: | Stubbs, H.E, Iverson, T.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tandem sialoglycan-binding modules in a Streptococcus sanguinis serine-rich repeat adhesin create target dependent avidity effects.
J.Biol.Chem., 295, 2020
|
|
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2QYF
 
 | |
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | Descriptor: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | Descriptor: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | Descriptor: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
8HL8
 
 | | Crystal structrue of MtdL R257K mutant | | Descriptor: | MANGANESE (II) ION, Transglycosylse | | Authors: | Li, F.D, He, C. | | Deposit date: | 2022-11-29 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the NDP-pyranose mutase belonging to glycosyltransferase family 75 reveal residues important for Mn 2+ coordination and substrate binding.
J.Biol.Chem., 299, 2023
|
|
5VYG
 
 | | Crystal structure of hFA9 EGF repeat with O-glucose trisaccharide | | Descriptor: | CALCIUM ION, Coagulation factor IX, alpha-D-xylopyranose-(1-3)-alpha-D-xylopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Yu, H.J, Li, H.L. | | Deposit date: | 2017-05-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | O-Glycosylation modulates the stability of epidermal growth factor-like repeats and thereby regulates Notch trafficking
J. Biol. Chem., 292, 2017
|
|
7MPJ
 
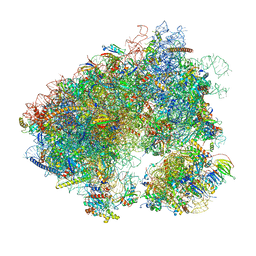 | | Stm1 bound vacant 80S structure isolated from wild-type | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Rai, J, Zhao, Y, Li, H. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
6IGC
 
 | | Crystal structure of HPV58/33/52 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IGD
 
 | | Crystal structure of HPV58/33 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
2Q6G
 
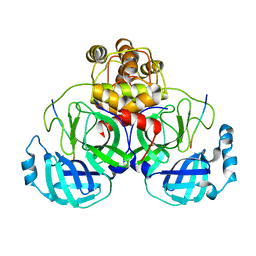 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6D
 
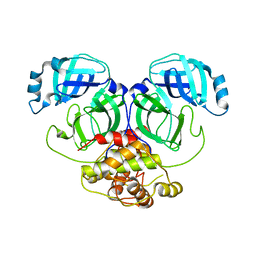 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
