1F2K
 
 | | CRYSTAL STRUCTURE OF ACANTHAMOEBA CASTELLANII PROFILIN II, CUBIC CRYSTAL FORM | | Descriptor: | PROFILIN II | | Authors: | Fedorov, A.A, Shi, W, Mahoney, N, Kaiser, D.A, Almo, S.C. | | Deposit date: | 2000-05-26 | | Release date: | 2000-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Comparative Structural Analysis of Profilins
To be Published
|
|
1INQ
 
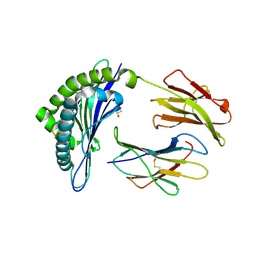 | | Structure of Minor Histocompatibility Antigen peptide, H13a, complexed to H2-Db | | Descriptor: | BETA-2 MICROGLOBULIN, DIMETHYL SULFOXIDE, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D, Nathenson, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2002-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
1JUF
 
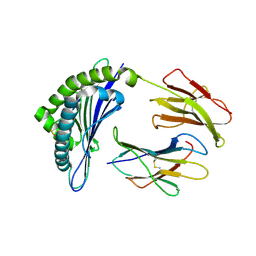 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | Descriptor: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | Deposit date: | 2001-08-24 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
2O3J
 
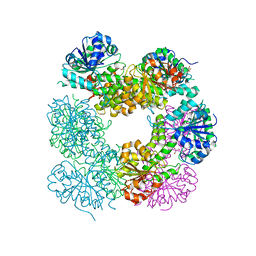 | | Structure of Caenorhabditis Elegans UDP-Glucose Dehydrogenase | | Descriptor: | GLYCEROL, UDP-glucose 6-dehydrogenase | | Authors: | Zhang, Y, Zhan, C, Patskovsky, Y, Ramagopal, U, Shi, W, Toro, R, Wengerter, B.C, Milst, S, Vidal, M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Caenorhabditis Elegans Udp-Glucose Dehydrogenase
To be Published
|
|
2PMB
 
 | | Crystal structure of predicted nucleotide-binding protein from Vibrio cholerae | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Zhan, C, Shi, W, Toro, R, Sauder, J.M, Gilmore, J, Iizuka, M, Maletic, M, Gheyi, T, Wasserman, S.R, Smith, D, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-20 | | Release date: | 2007-05-08 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Predicted Nucleotide-Binding Protein from Vibrio Cholerae.
To be Published
|
|
1QXH
 
 | |
4W7H
 
 | | Crystal Structure of DEH Reductase A1-R Mutant | | Descriptor: | Carbonyl reductase | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4W7I
 
 | | Crystal structure of DEH reductase A1-R' mutant | | Descriptor: | 4-deoxy-L-erythro-5-hexoseulose uronate reductase A1-R' | | Authors: | Takase, R, Mikami, B, Kawai, S, Murata, K, Hashimoto, W. | | Deposit date: | 2014-08-22 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-based Conversion of the Coenzyme Requirement of a Short-chain Dehydrogenase/Reductase Involved in Bacterial Alginate Metabolism.
J.Biol.Chem., 289, 2014
|
|
4XTC
 
 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | Descriptor: | AlgM1, AlgM2, AlgQ2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
1RT8
 
 | | CRYSTAL STRUCTURE OF THE ACTIN-CROSSLINKING CORE OF SCHIZOSACCHAROMYCES POMBE FIMBRIN | | Descriptor: | SULFATE ION, fimbrin | | Authors: | Klein, M.G, Shi, W, Ramagopal, U, Tseng, Y, Wirtz, D, Kovar, D.R, Staiger, C.J, Almo, S.C. | | Deposit date: | 2003-12-10 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the actin crosslinking core of fimbrin.
Structure, 12, 2004
|
|
4XM0
 
 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the absence of cadmium | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM2
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus in the absence of cadmium | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XLZ
 
 | | N,N'-diacetylchitobiose deacetylase (SeMet derivative) from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM1
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus in the presence of cadmium | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nakamura, T, Niiyama, M, Hashimoto, W, Ida, K, Uegaki, K. | | Deposit date: | 2015-01-14 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple crystal forms of N,N'-diacetylchitobiose deacetylase from Pyrococcus furiosus.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XIG
 
 | | Crystal structure of bacterial alginate ABC transporter determined through humid air and glue-coating method | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
5XS8
 
 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with two sulfate groups at C-4 and C-6 positions of GalNAc | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | Authors: | Oiki, S, Kamochi, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2017-06-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Alternative substrate-bound conformation of bacterial solute-binding protein involved in the import of mammalian host glycosaminoglycans.
Sci Rep, 7, 2017
|
|
7JYC
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K3T
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) at 1.2 A Resolution and a Possible Capture of Zinc Binding Intermediate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-13 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6D
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.48 A Resolution (Cryo-protected) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K40
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Boceprevir at 1.35 A Resolution | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, boceprevir (bound form) | | Authors: | Kumaran, D, Andi, B, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K6E
 
 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7YTB
 
 | | Crystal structure of Kin4B8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Kin4B8, RETINAL | | Authors: | Murakoshi, S, Chazan, A, Shihoya, W, Beja, O, Nureki, O. | | Deposit date: | 2022-08-14 | | Release date: | 2023-03-15 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phototrophy by antenna-containing rhodopsin pumps in aquatic environments.
Nature, 615, 2023
|
|
6INZ
 
 | | Crystal structure of solute-binding protein complexed with unsaturated hyaluronan disaccharide | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular solute-binding protein family 1, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Substrate recognition by bacterial solute-binding protein is responsible for import of extracellular hyaluronan and chondroitin sulfate from the animal host.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
1XM5
 
 | | Crystal structure of metal-dependent hydrolase ybeY from E. coli, Pfam UPF0054 | | Descriptor: | Hypothetical UPF0054 protein ybeY, NICKEL (II) ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Shi, W, Ramagopal, U.A, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The ybeY protein from Escherichia coli is a metalloprotein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2A0Y
 
 | | Structure of human purine nucleoside phosphorylase H257D mutant | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Murkin, A.S, Shi, W, Schramm, V.L. | | Deposit date: | 2005-06-17 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
