3O17
 
 | | Crystal Structure of JNK1-alpha1 isoform | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, JIP1, 10MER PEPTIDE, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2010-07-20 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of JNK1-alpha1 isoform
TO BE PUBLISHED
|
|
3O2M
 
 | |
2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6WHC
 
 | | CryoEM Structure of the glucagon receptor with a dual-agonist peptide | | Descriptor: | Dual-agonist peptide, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Sexton, P, Danev, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.
J.Biol.Chem., 295, 2020
|
|
1COL
 
 | | REFINED STRUCTURE OF THE PORE-FORMING DOMAIN OF COLICIN A AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | COLICIN A | | Authors: | Parker, M.W, Postma, J.P.M, Pattus, F, Tucker, A.D, Tsernoglou, D. | | Deposit date: | 1991-07-06 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined structure of the pore-forming domain of colicin A at 2.4 A resolution.
J.Mol.Biol., 224, 1992
|
|
2RFX
 
 | | Crystal Structure of HLA-B*5701, presenting the self peptide, LSSPVTKSF | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Archbold, J.K, Macdonald, W.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity
Immunity, 28, 2008
|
|
4B00
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND (R)-41) | | Descriptor: | 5-{(1R)-3-amino-4-fluoro-1-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-1H-isoindol-1-yl}-1-ethyl-3-methylpyridin-2(1H)-one, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
5J4L
 
 | | Apo-structure of humanised RadA-mutant humRadA22F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JEE
 
 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
4AZY
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND 10) | | Descriptor: | (1S)-4-fluoro-1-(4-fluoro-3-pyrimidin-5-ylphenyl)-1-[2-(trifluoromethyl)pyridin-4-yl]-1H-isoindol-3-amine, ACETATE ION, BETA-SECRETASE 1, ... | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5L8V
 
 | | Apo-structure of humanised RadA-mutant humRadA4 | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB4
 
 | | Apo-structure of humanised RadA-mutant humRadA14 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LBI
 
 | | Apo-structure of humanised RadA-mutant humRadA3 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5A9H
 
 | | Crystal structure of the extracellular domain of PepT2 | | Descriptor: | ACETATE ION, CESIUM ION, GLYCEROL, ... | | Authors: | Beale, J.H, Newstead, S. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
5A9D
 
 | | Crystal structure of the extracellular domain of PepT1 | | Descriptor: | GLYCEROL, SOLUTE CARRIER FAMILY 15 MEMBER 1 | | Authors: | Beale, J.H, Bird, L.E, Owens, R.J, Newstead, S. | | Deposit date: | 2015-07-20 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
5LB2
 
 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5A9I
 
 | | Crystal structure of the extracellular domain of PepT2 | | Descriptor: | CITRIC ACID, GLYCEROL, SOLUTE CARRIER FAMILY 15 MEMBER 2 | | Authors: | Beale, J.H, Newstead, S. | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structures of the Extracellular Domain from Pept1 and Pept2 Provide Novel Insights Into Mammalian Peptide Transport
Structure, 23, 2015
|
|
1JV2
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR SEGMENT OF INTEGRIN ALPHAVBETA3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Diefenbach, B, Zhang, R, Dunker, R, Scott, D, Joachimiak, A, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the extracellular segment of integrin alpha Vbeta3.
Science, 294, 2001
|
|
6QG9
 
 | | Crystal structure of Ideonella sakaiensis MHETase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGC
 
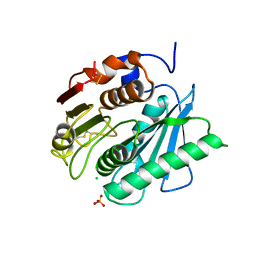 | | PETase from Ideonella sakaiensis without ligand | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGA
 
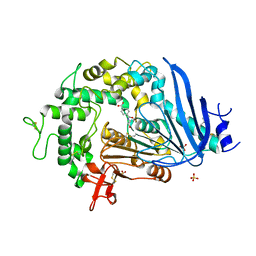 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGB
 
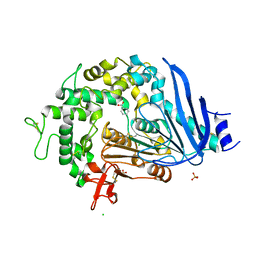 | | Crystal structure of Ideonella sakaiensis MHETase bound to benzoic acid | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | Authors: | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
9FHE
 
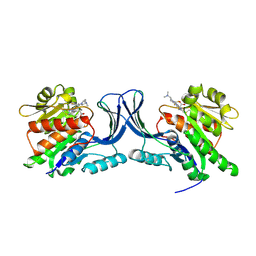 | | hKHK-C in complex with BI-9787 (pH 5.5) | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
9FHD
 
 | | hKHK-C in fomplex with BI-9787 | | Descriptor: | (2~{S})-3-[3-[[4-[bis(fluoranyl)methyl]-3-cyano-6-[(3~{S})-3-(dimethylamino)pyrrolidin-1-yl]pyridin-2-yl]amino]-4-methylsulfanyl-phenyl]-2-methyl-propanoic acid, Ketohexokinase, SULFATE ION | | Authors: | Ebenhoch, R, Pautsch, A. | | Deposit date: | 2024-05-27 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Discovery of BI-9787, a potent zwitterionic ketohexokinase inhibitor with oral bioavailability.
Bioorg.Med.Chem.Lett., 112, 2024
|
|
