9AYG
 
 | | Cryo-EM structure of apo state human Cav3.2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYH
 
 | | Cryo-EM structure of human Cav3.2 with TTA-A2 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(4-cyclopropylphenyl)-N-{(1R)-1-[5-(2,2,2-trifluoroethoxy)pyridin-2-yl]ethyl}acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
9AYL
 
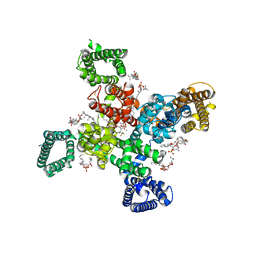 | | Cryo-EM structure of human Cav3.2 with ACT-709478 | | Descriptor: | 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2024-03-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human Ca v 3.2 inhibition by selective antagonists.
Cell Res., 34, 2024
|
|
8FHD
 
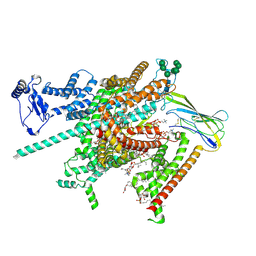 | | Cryo-EM structure of human voltage-gated sodium channel Nav1.6 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (5E,17R,20S)-23-amino-20-hydroxy-14,20-dioxo-15,19,21-trioxa-20lambda~5~-phosphatricos-5-en-17-yl hexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8YJO
 
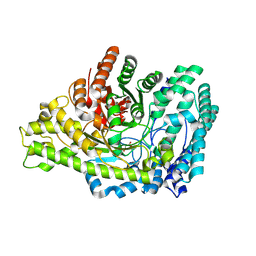 | | Structure of E. coli glycyl radical enzyme PflD with bound malonate | | Descriptor: | MALONATE ION, Probable dehydratase PflD | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
8YJN
 
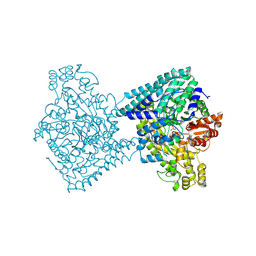 | | Structure of E. coli glycyl radical enzyme YbiW with bound glycerol | | Descriptor: | GLYCEROL, Probable dehydratase YbiW | | Authors: | Xue, B, Wei, Y, Robinson, R.C, Yew, W.S, Zhang, Y. | | Deposit date: | 2024-03-02 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | A Widespread Radical-Mediated Glycolysis Pathway.
J.Am.Chem.Soc., 146, 2024
|
|
6LQA
 
 | | voltage-gated sodium channel Nav1.5 with quinidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Quinidine, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Li, Z, Pan, X, Huang, G. | | Deposit date: | 2020-01-13 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Na v 1.5 by the Antiarrhythmic Drug Quinidine*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6LT8
 
 | | HSP90 in complex with KW-2478 | | Descriptor: | 2-[2-ethyl-6-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]carbonyl-3,5-bis(oxidanyl)phenyl]-~{N},~{N}-bis(2-methoxyethyl)ethanamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Anti-NSCLC activity in vitro of Hsp90 N inhibitor KW-2478 and complex crystal structure determination of Hsp90 N -KW-2478.
J.Struct.Biol., 213, 2021
|
|
5GPP
 
 | | Crystal structure of zebrafish ASC PYD domain | | Descriptor: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
6LTK
 
 | | HSP90 in complex with SNX-2112 | | Descriptor: | 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Complex Crystal Structure Determination and in vitro Anti-non-small Cell Lung Cancer Activity of Hsp90 N Inhibitor SNX-2112.
Front Cell Dev Biol, 9, 2021
|
|
6LR9
 
 | | HSP90 in complex with Debio0932 | | Descriptor: | 2-[[6-(dimethylamino)-1,3-benzodioxol-5-yl]sulfanyl]-1-[2-(2,2-dimethylpropylamino)ethyl]imidazo[4,5-c]pyridin-4-amine, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Complex crystal structure determination and anti-non-small-cell lung cancer activity of the Hsp90 N inhibitor Debio0932.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5GPQ
 
 | | Crystal Structure of zebrafish ASC CARD Domain | | Descriptor: | CITRIC ACID, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
7XVE
 
 | | Human Nav1.7 mutant class-I | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XVF
 
 | | Nav1.7 mutant class2 | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Huang, G, Wu, Q, Li, Z, Pan, X, Yan, N. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unwinding and spiral sliding of S4 and domain rotation of VSD during the electromechanical coupling in Na v 1.7.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5GP1
 
 | | Crystal structure of ZIKV NS5 Methyltransferase in complex with GTP and SAH | | Descriptor: | NICKEL (II) ION, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-directed RNA polymerase NS5, ... | | Authors: | Zhang, C, Jin, T. | | Deposit date: | 2016-07-30 | | Release date: | 2016-12-07 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design
Biochem. Biophys. Res. Commun., 492, 2017
|
|
5GOZ
 
 | |
6ON7
 
 | |
3R24
 
 | | Crystal structure of nsp10/nsp16 complex of SARS coronavirus | | Descriptor: | 2'-O-methyl transferase, Non-structural protein 10 and Non-structural protein 11, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, X, Guo, D, Su, C, Chen, Y. | | Deposit date: | 2011-03-13 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex.
Plos Pathog., 7, 2011
|
|
2DK9
 
 | | Solution structure of Calponin Homology domain of Human MICAL-1 | | Descriptor: | NEDD9-interacting protein with calponin homology and LIM domains | | Authors: | Sun, H, Dai, H, Zhang, J, Xiong, S, Wu, J, Shi, Y. | | Deposit date: | 2006-04-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calponin homology domain of Human MICAL-1
J.Biomol.Nmr, 36, 2006
|
|
6MCU
 
 | |
6MCV
 
 | |
6MLB
 
 | |
6MKV
 
 | |
6ON5
 
 | |
6ON8
 
 | |
