6W3F
 
 | |
6W40
 
 | |
7TXZ
 
 | |
7TY0
 
 | |
7TJL
 
 | |
6MRR
 
 | | De novo designed protein Foldit1 | | Descriptor: | Foldit1 | | Authors: | Koepnick, B, Bick, M.J, Estep, R.D, Kleinfelter, S, Wei, L, Baker, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-06-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
6MRS
 
 | |
6MSP
 
 | | De novo Designed Protein Foldit3 | | Descriptor: | De novo Designed Protein Foldit3 | | Authors: | Liu, G, Ishida, Y, Swapna, G.V.T, Kleinfelter, S, Koepnick, B, Baker, D, Montelione, G.T. | | Deposit date: | 2018-10-17 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo protein design by citizen scientists.
Nature, 570, 2019
|
|
4KYZ
 
 | | Three-dimensional structure of triclinic form of de novo design insertion domain, Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | Designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
4MEE
 
 | | Crystal structure of the transport unit of the autotransporter AIDA-I from Escherichia coli | | Descriptor: | Diffuse adherence adhesin | | Authors: | Gawarzewski, I, Tschapek, B, Hoeppner, A, Smits, S.H, Jose, J, Schmitt, L. | | Deposit date: | 2013-08-26 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transport unit of the autotransporter adhesin involved in diffuse adherence from Escherichia coli.
J.Struct.Biol., 187, 2014
|
|
4L11
 
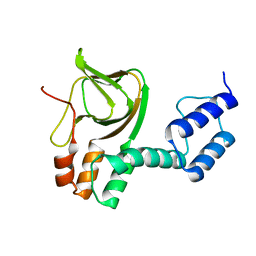 | | Structure of the C-linker/CNBHD of agERG channels | | Descriptor: | AGAP007709-PA | | Authors: | Brelidze, T.I. | | Deposit date: | 2013-06-01 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the C-terminal region of an ERG channel and functional implications.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KY3
 
 | | Three-dimensional Structure of the orthorhombic crystal of computationally designed insertion domain , Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
6DMA
 
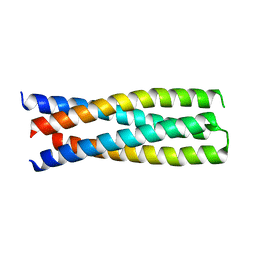 | | DHD15_closed | | Descriptor: | DHD15_closed_A, DHD15_closed_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.363 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DKM
 
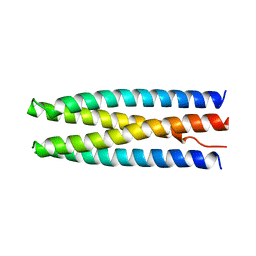 | | DHD131 | | Descriptor: | DHD131_A, DHD131_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DMP
 
 | | De Novo Design of a Protein Heterodimer with Specificity Mediated by Hydrogen Bond Networks | | Descriptor: | Designed orthogonal protein DHD13_XAAA_A, Designed orthogonal protein DHD13_XAAA_B | | Authors: | Chen, Z, Flores-Solis, D, Sgourakis, N.G, Baker, D. | | Deposit date: | 2018-06-05 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DLM
 
 | | DHD127 | | Descriptor: | DHD127_A, DHD127_B | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-01 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DM9
 
 | | DHD15_extended | | Descriptor: | DHD15_extended_A, DHD15_extended_B, SULFATE ION | | Authors: | Bick, M.J, Chen, Z, Baker, D. | | Deposit date: | 2018-06-04 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Programmable design of orthogonal protein heterodimers.
Nature, 565, 2019
|
|
6DLC
 
 | |
3QD9
 
 | |
3T58
 
 | |
3T59
 
 | |
3QCP
 
 | | QSOX from Trypanosoma brucei | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QSOX from Trypanosoma brucei (TbQSOX) | | Authors: | Alon, A, Fass, D. | | Deposit date: | 2011-01-17 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The dynamic disulphide relay of quiescin sulphydryl oxidase.
Nature, 488, 2012
|
|
5V7V
 
 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
5V6P
 
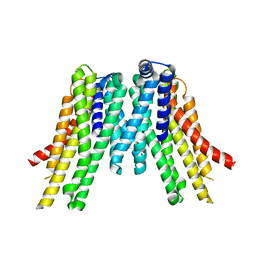 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Authors: | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
6BAA
 
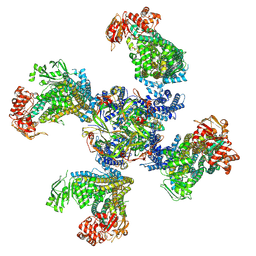 | | Cryo-EM structure of the pancreatic beta-cell KATP channel bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Martin, G.M, Yoshioka, C, Shyng, S.L. | | Deposit date: | 2017-10-12 | | Release date: | 2017-11-01 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Elife, 6, 2017
|
|
